Open research software infrastructure
in Neuro-Medicine
| Adina Wagner
|
|
|
Psychoinformatics lab,
Institute of Neuroscience and Medicine, Brain & Behavior (INM-7) Research Center Jülich |
 |

Slides: DOI 10.5281/zenodo.10149349 (Scan the QR code)
files.inm7.de/adina/talks/html/zimannheim.html
Open Science and Open Software go hand in hand
- Science has specific requirements; research software from within science ("from scientists, for scientists") can fulfill them. Open formats, protocols, and code allow re-use, interoperability, and customization.
- Open and reproducible science has specific needs for transparency and accessibility: Open source software provides the necessary auditability.
- Creating software becomes increasingly possible for scientists: The San Francisco Declaration on Research Assessment (DORA; signed by FZJ), the Agreement on Reforming Research Assessment (CoARA), and the DFG recognize software as academic output.
The Institute for Neuroscience and Medicine (INM-7)
The Institute for Neuroscience and Medicine (INM-7)
- Interdisciplinary institute with 11 research groups
- Research foci:
- Infrastructure and method development: Digital biomarker, machine learning, meta analysis, research data management
- Basic research in human brain mapping: Connectomics, genetic gradients, in-vivo brain mapping, multimodal integration
- AI Applications in medical research: Cognition, Personality, Aging & neurodegenerative disease, Schizophrenia
- Ethical implications of medical AI: Bias in AI applications, medical AI and society, individualized predictions
Software @ INM-7
- The institute has a history of open source software, starting with the SPM Anatomy Toolbox (Eickhoff, 2005)
- Multiple groups develop and maintain open source research software for their respecitve subdomain
- Recent integration efforts connect our open software stack to open research software infrastructure for neuro-medicine
- Domain-agnostic data management tool (command-line + graphical user interface), built on top of Git & Git-annex
- 10+ year open source project (100+ contributors), available for all major OS
- Born from rethinking data:
- Just like code, data is not static.
- Just like code, data is subject to collaboration. Stream-lined workflows for sharing and collaborating should be possible, mirroring those in software development.
- Provenance of data is essential for reproducible, trustworthy, and FAIR science
- Flexibility and interoperability with existing tools is the key to sustainability and ease of use
- Domain-agnostic command-line tool (+ graphical user interface), built on top of Git & Git-annex
- 10+ year open source project (100+ contributors), available for all major OS
- Major features:
- Version-controlling arbitrarily large content
- Version control data & software alongside to code!
- Transport mechanisms for sharing, updating & obtaining data
- Consume & collaborate on data (analyses) like software
- (Computationally) reproducible data analysis
- Track and share provenance of all digital objects
- (... and much more)
DataLad usecases

Acknowledgements
|
Funders







Collaborators







|
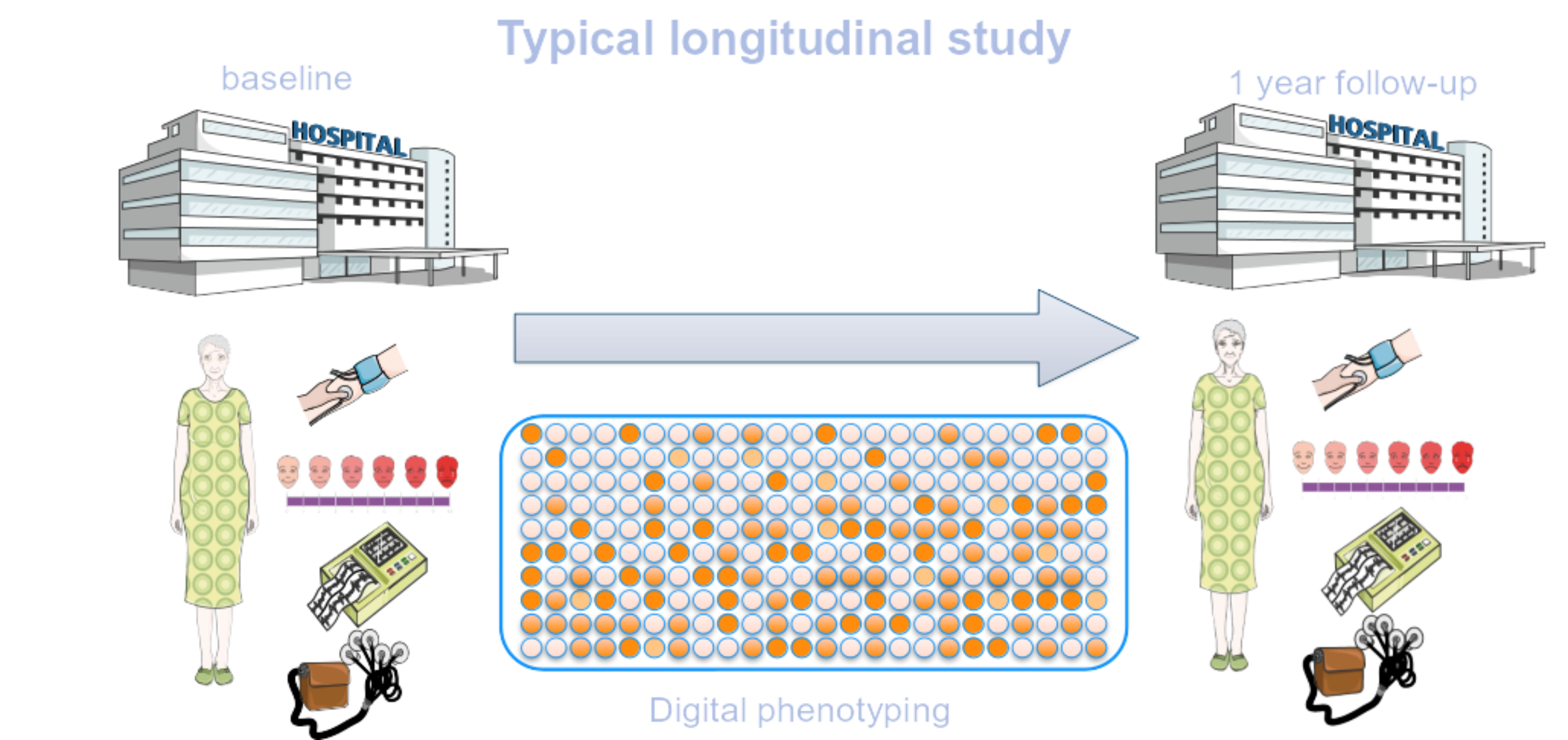
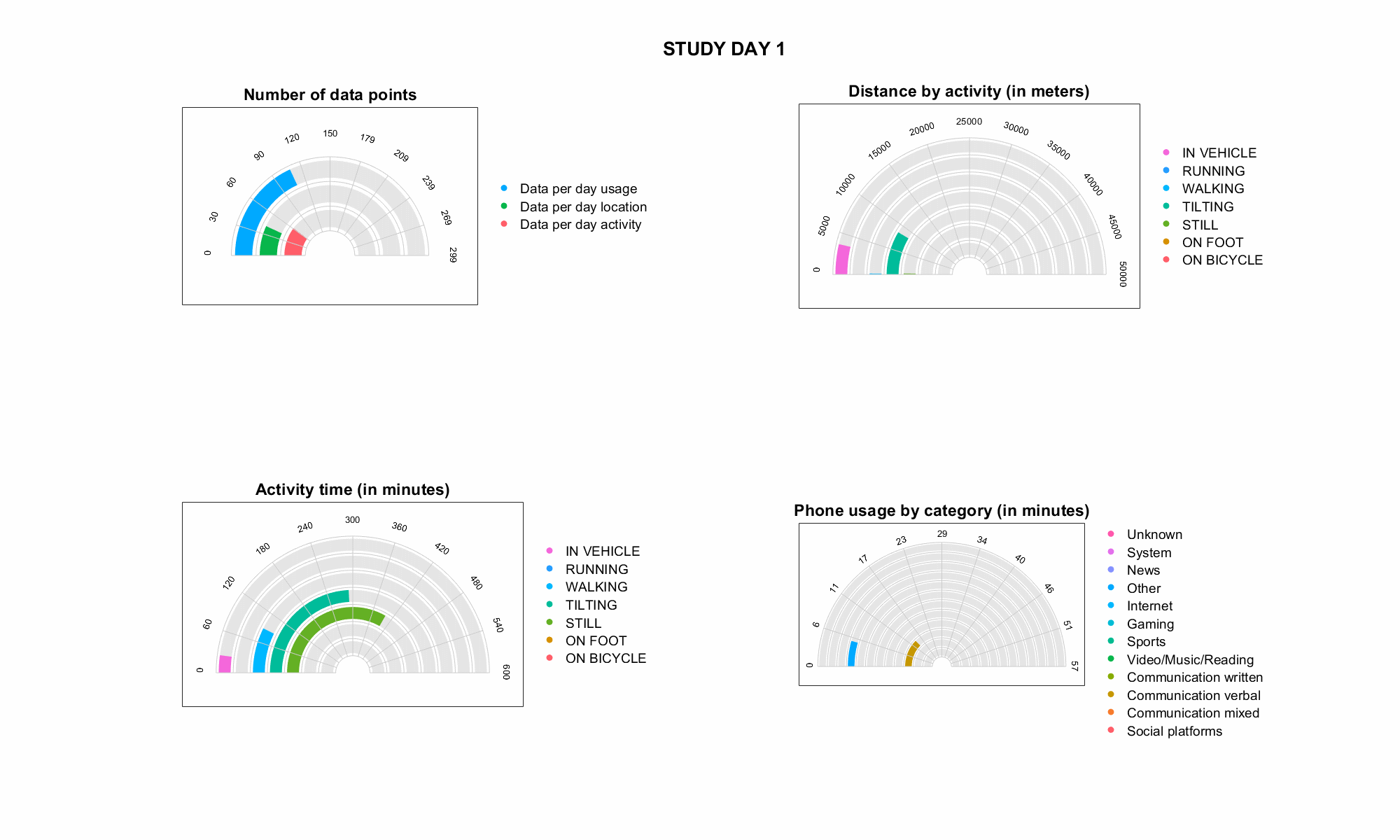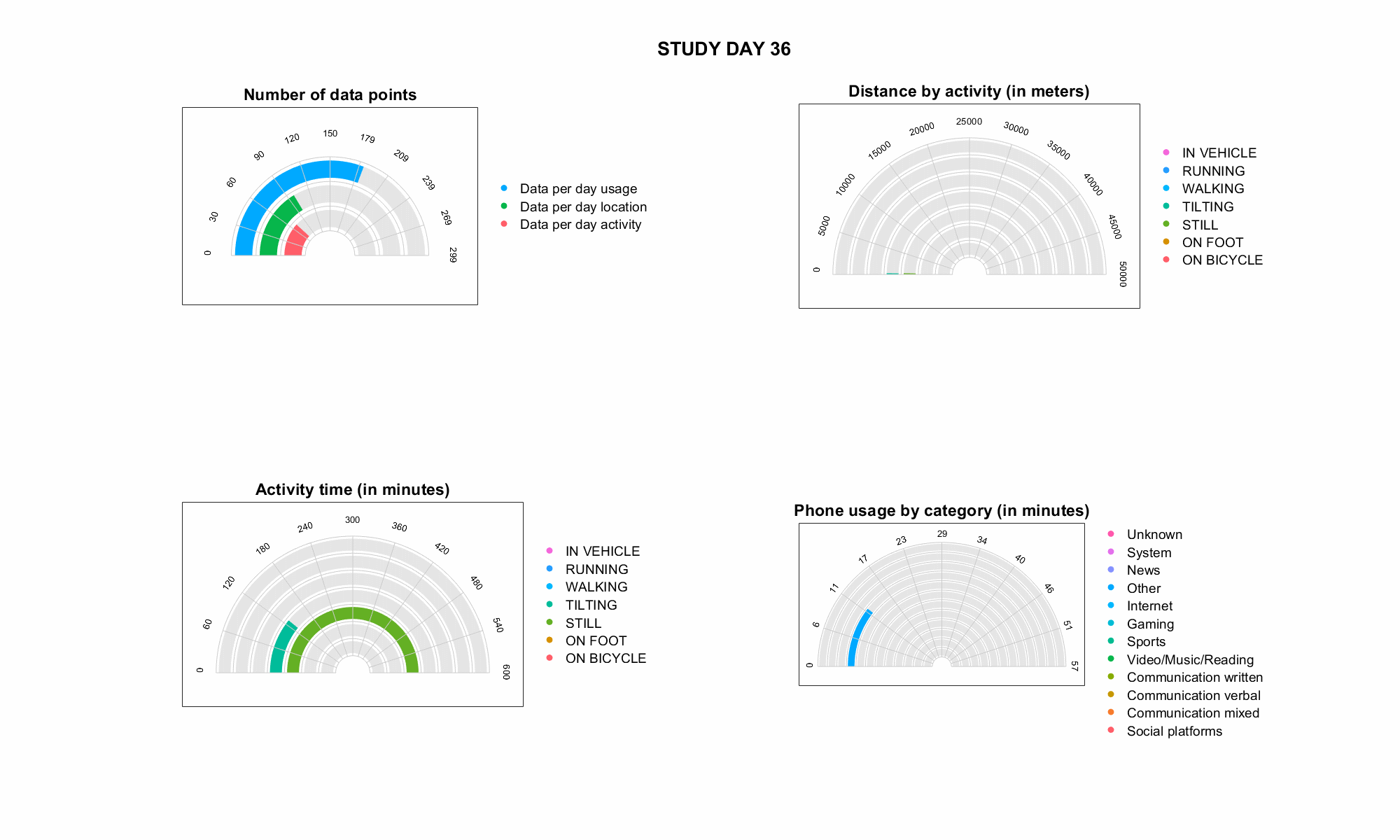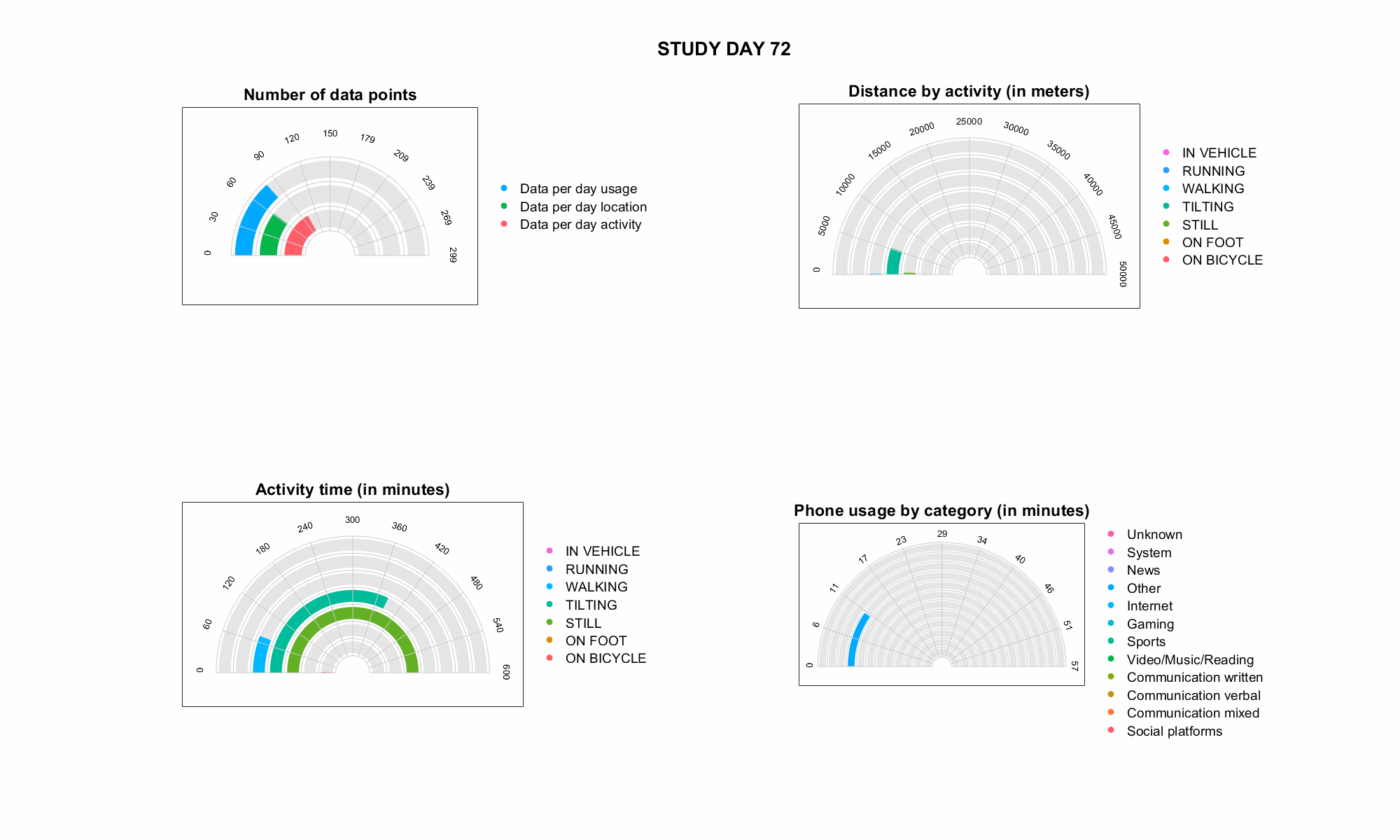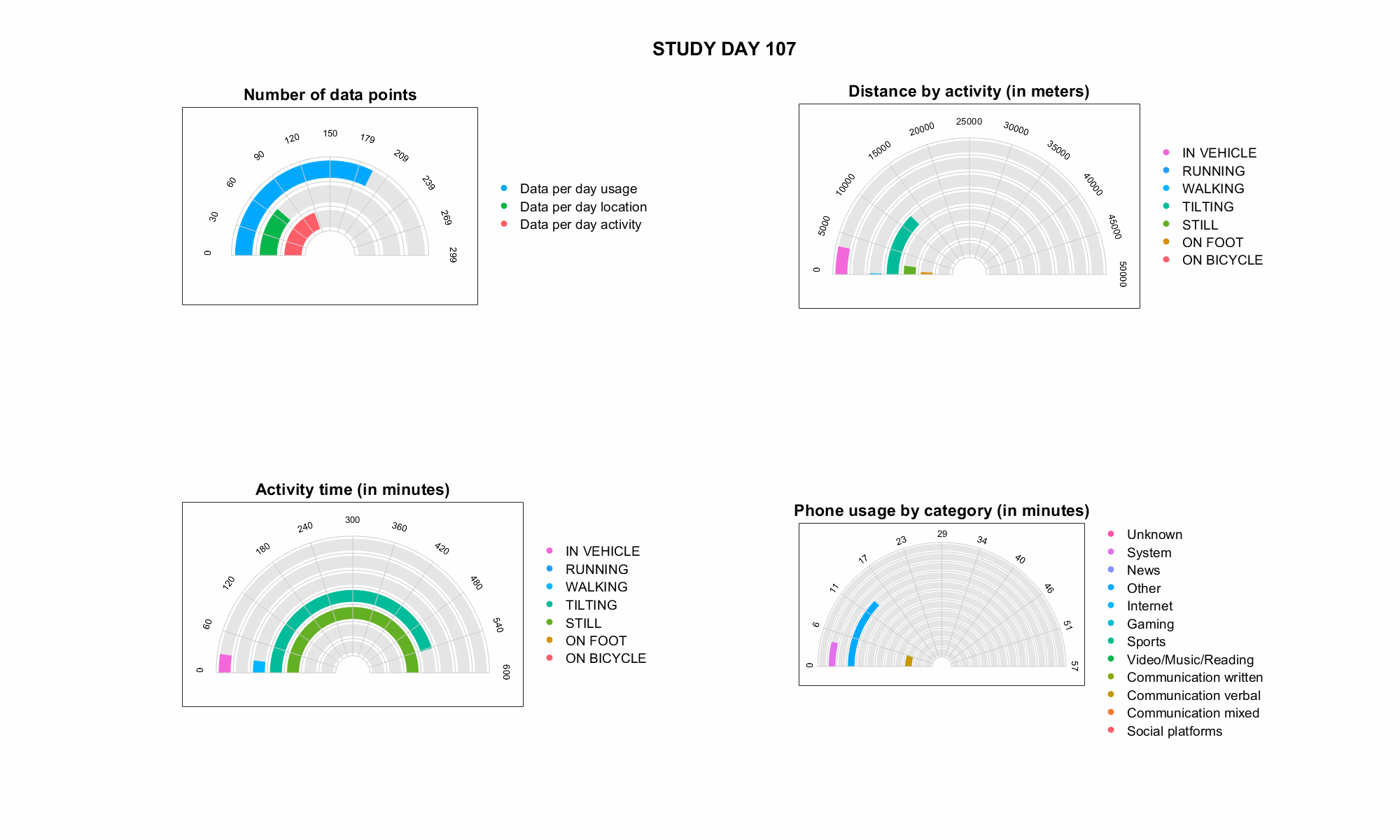
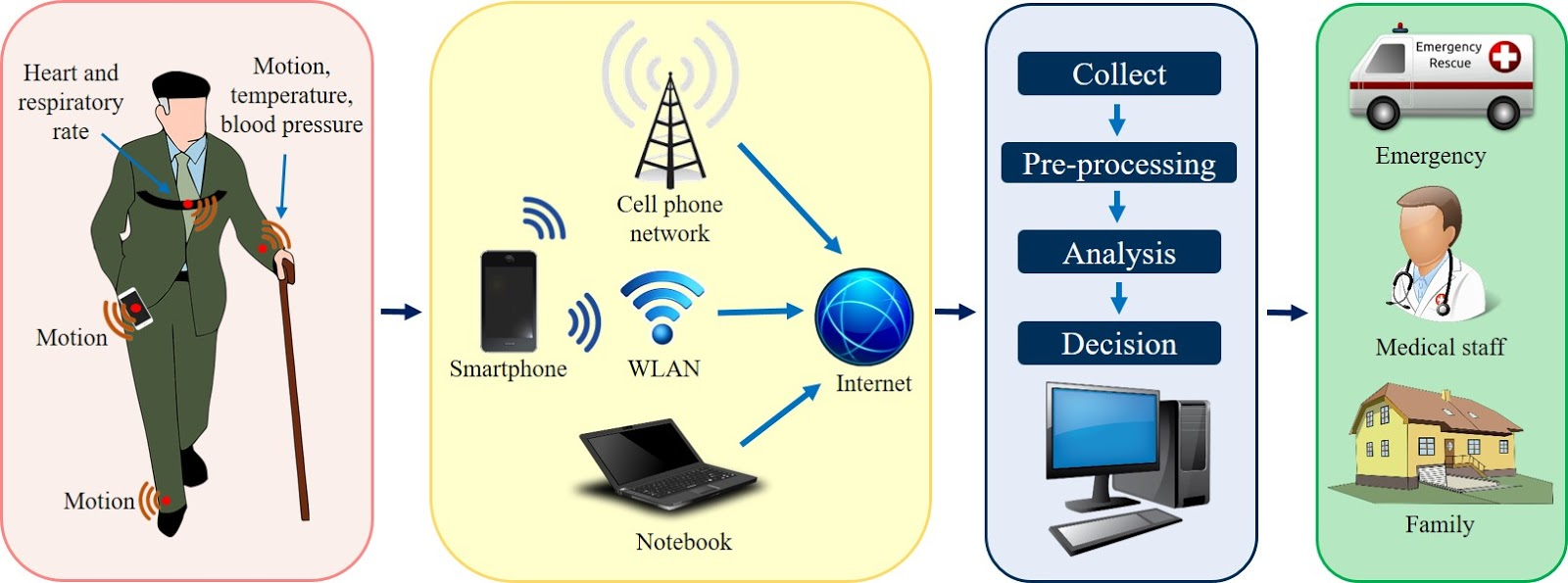
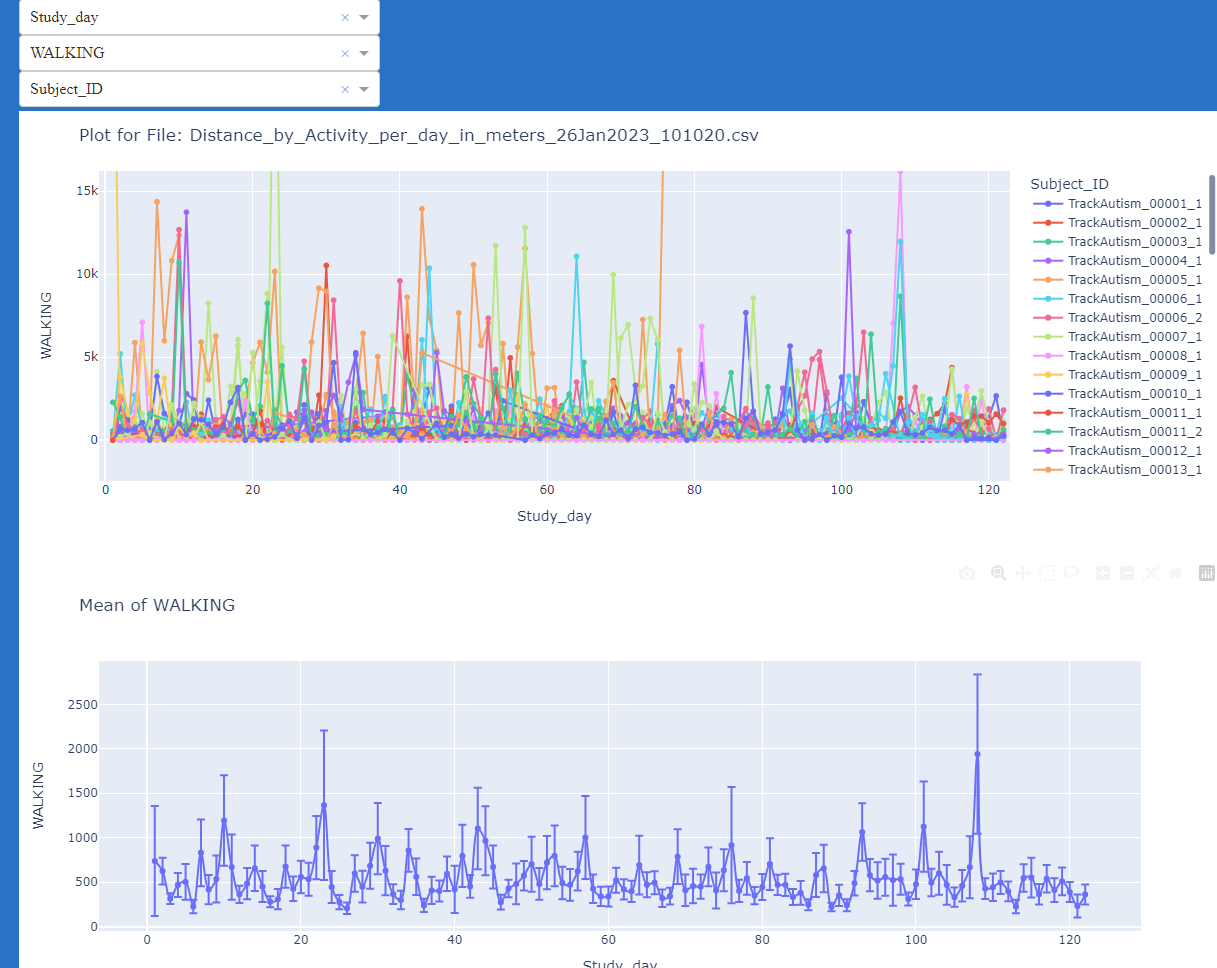
JTrack: Digital biomarkers from your smartphone
- Objective: Close monitoring of patients/participants in non-clinical settings
- Modern smartphones contain a variety of sensors for passive monitoring and active acquisition:
- gyroscope
- accelerometer
- location
- human activity recognition
- application usage
- screen time
- microphone

|
|
JTrack
- Flexible components for different users:
- JTrack Social: Smartphone app for participants
- JTrack EMA: Smartphone app for participants
- JDash: Monitoring and analytics tool for study owners
JTrack components: JTrack Social
-
Smartphone App for active labeling and passive monitoring
- Sensor data (passive
collection mode default:
Accelerometer & Gyroscope) - Application usage statistics
- Human activaty recognition
(e.g., walking, running, driving) - Location information (anonymized)
- Active recording, e.g., free-speech
generation tasks
|
|

|
JTrack components: JTrack Social
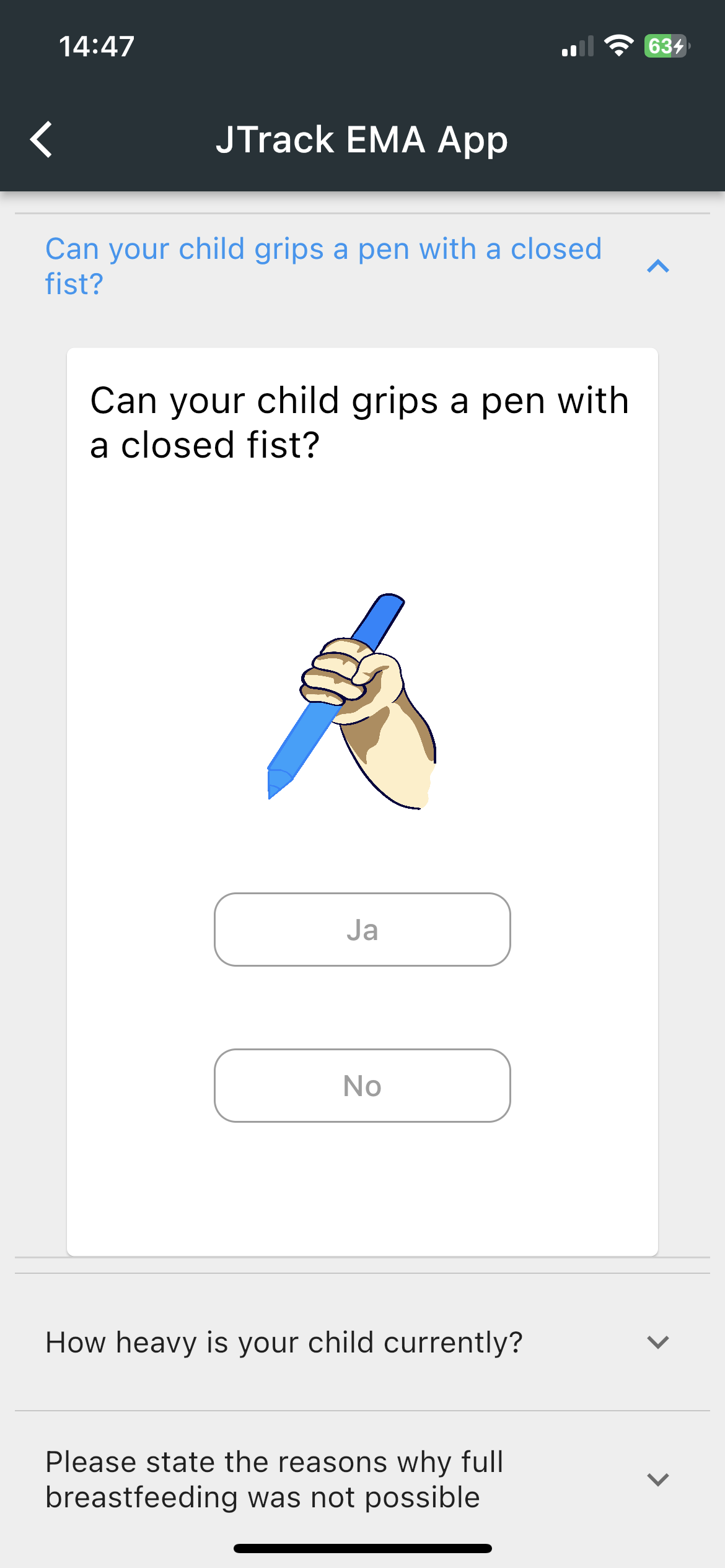
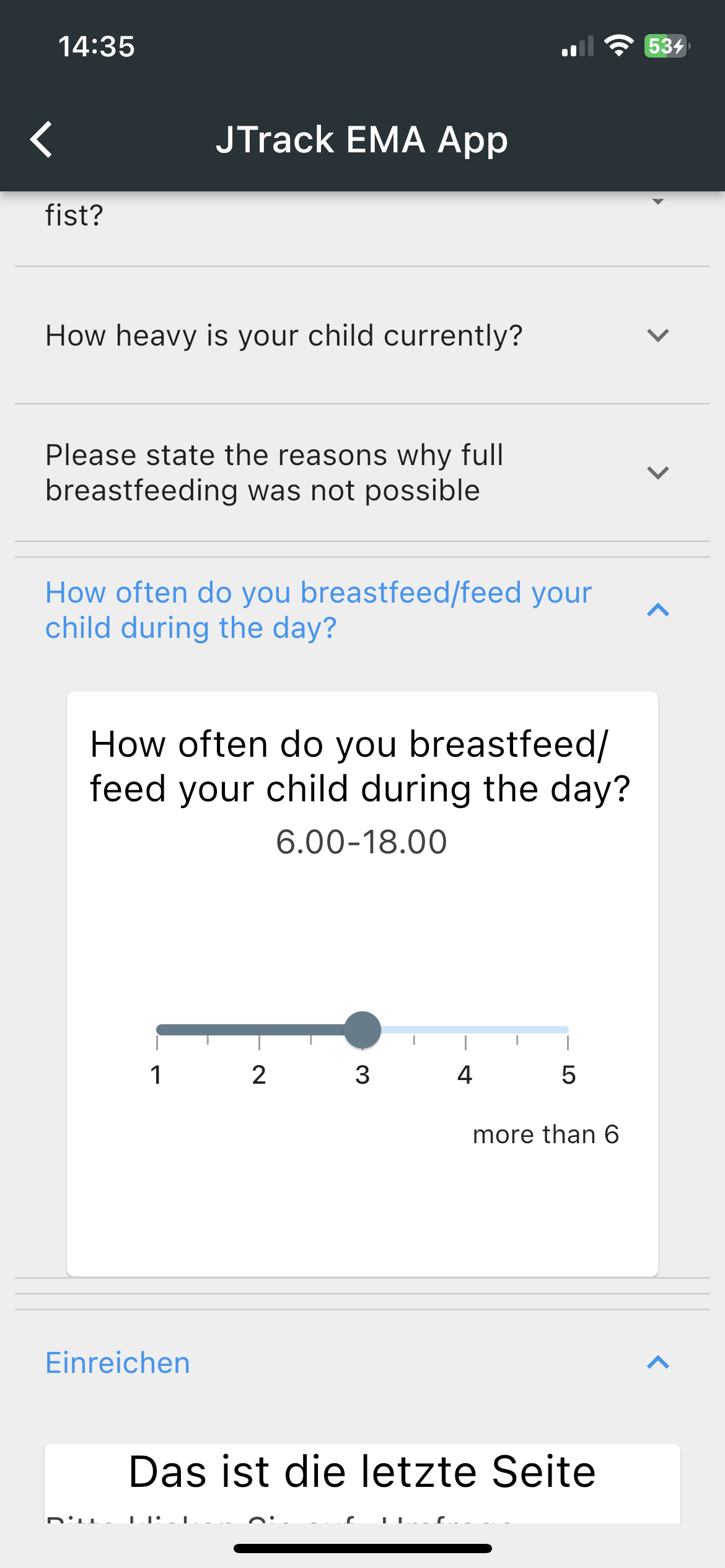
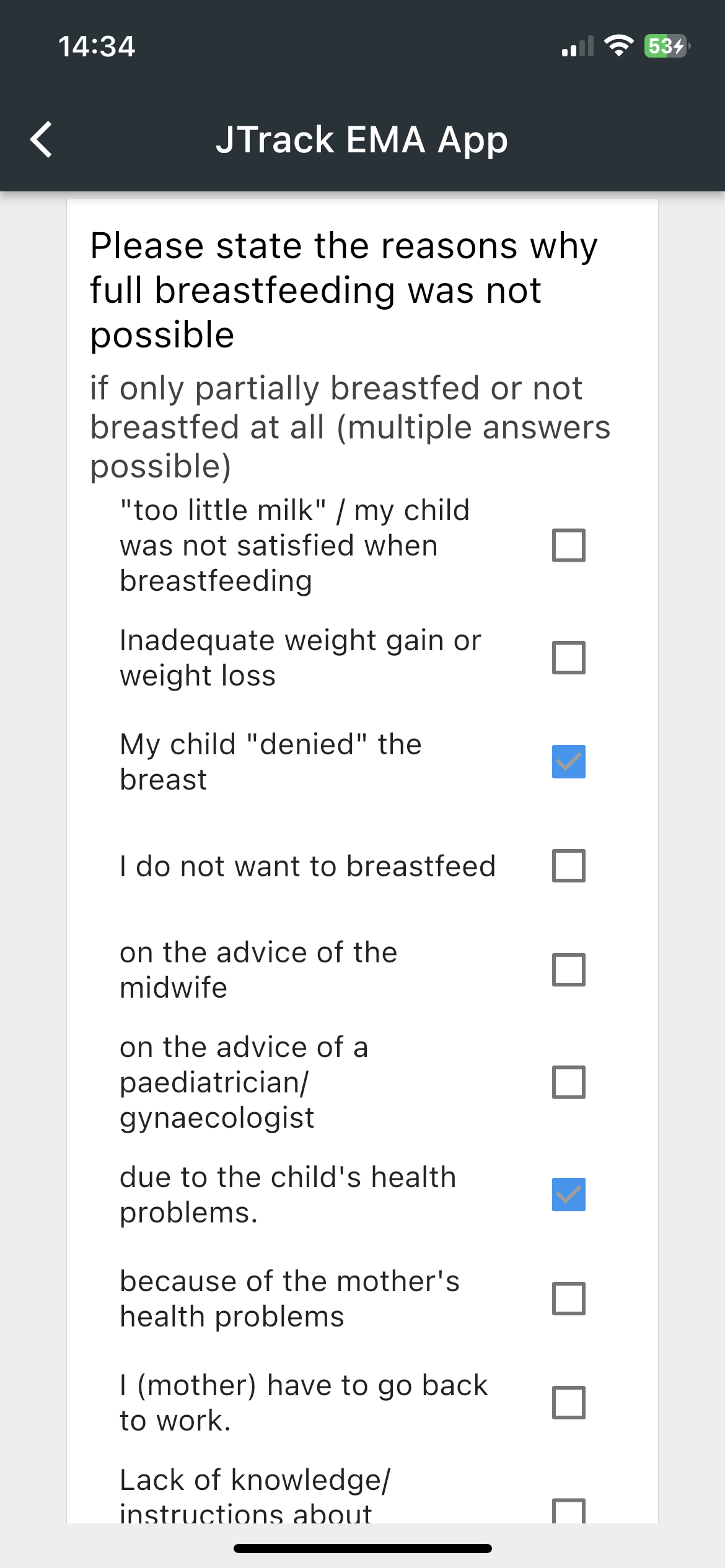
JTrack components: JTrack EMA
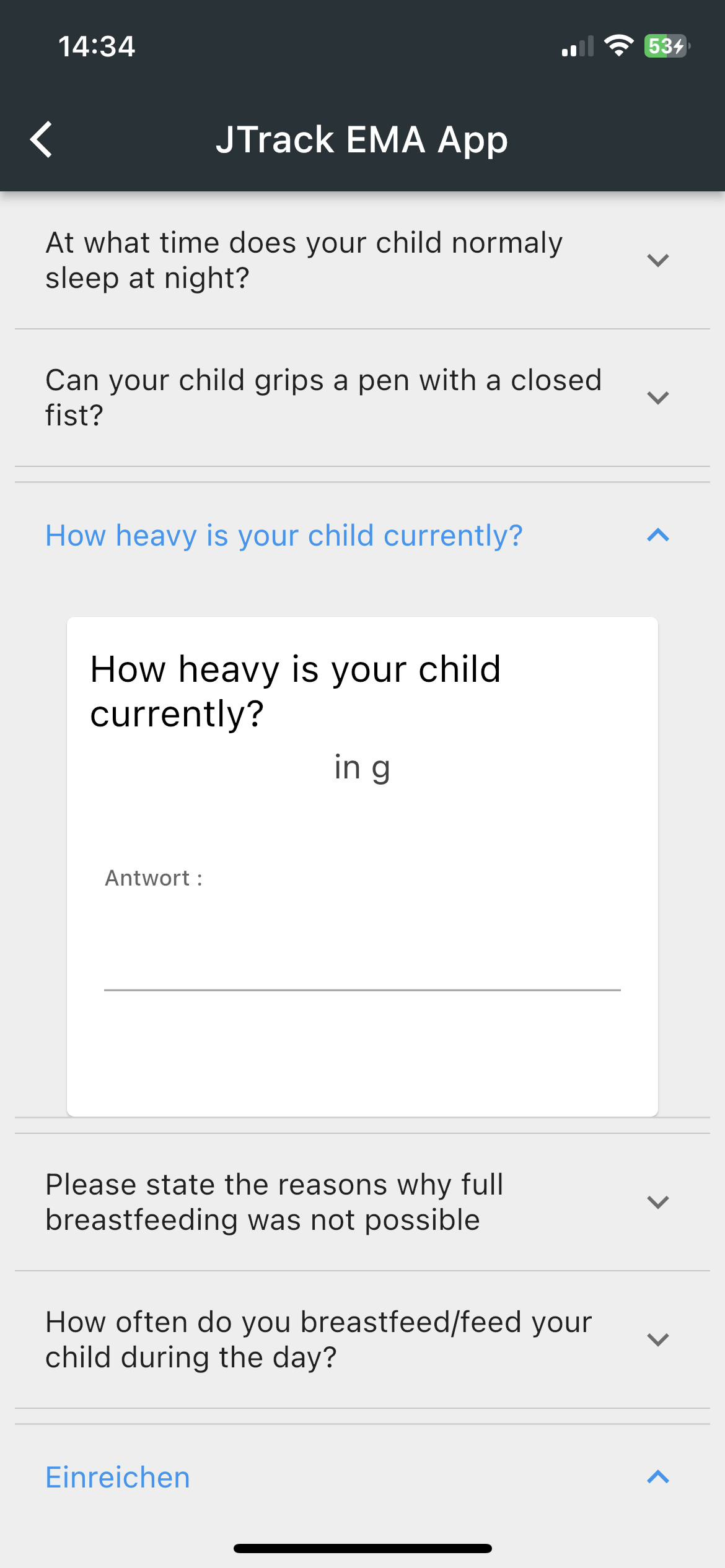
-
Smartphone App for Ecological Momentary Assessment
- Binary Questions
- Date and Time
Questions - Sliding Questions
- Multiple/Single
coice questions
|
|

|
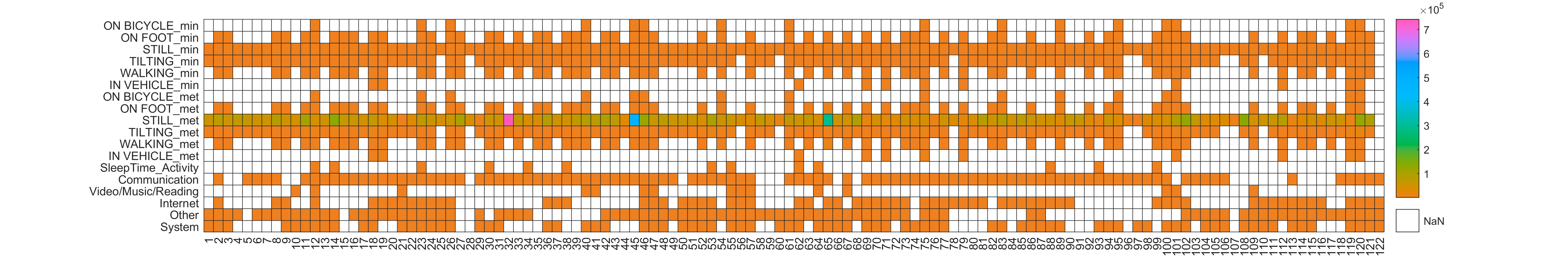
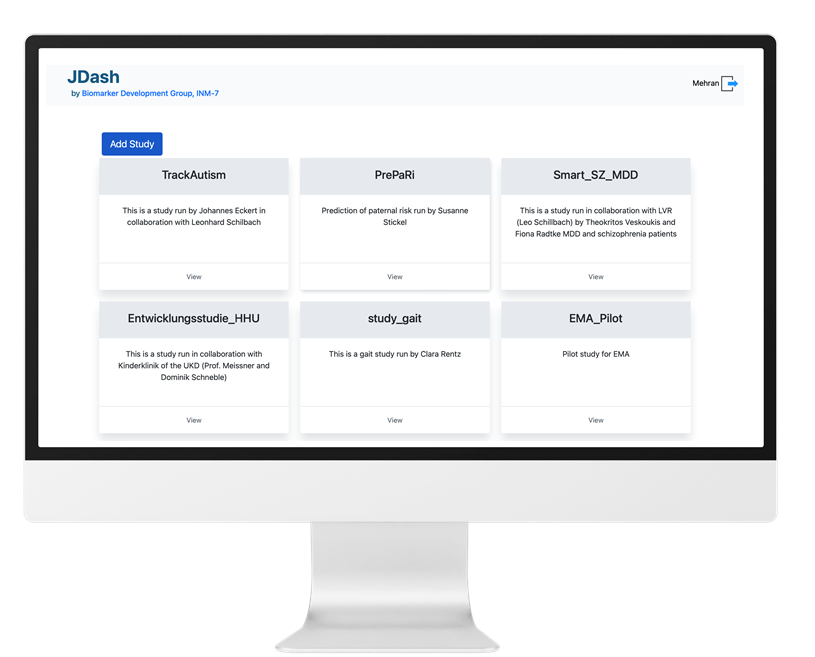
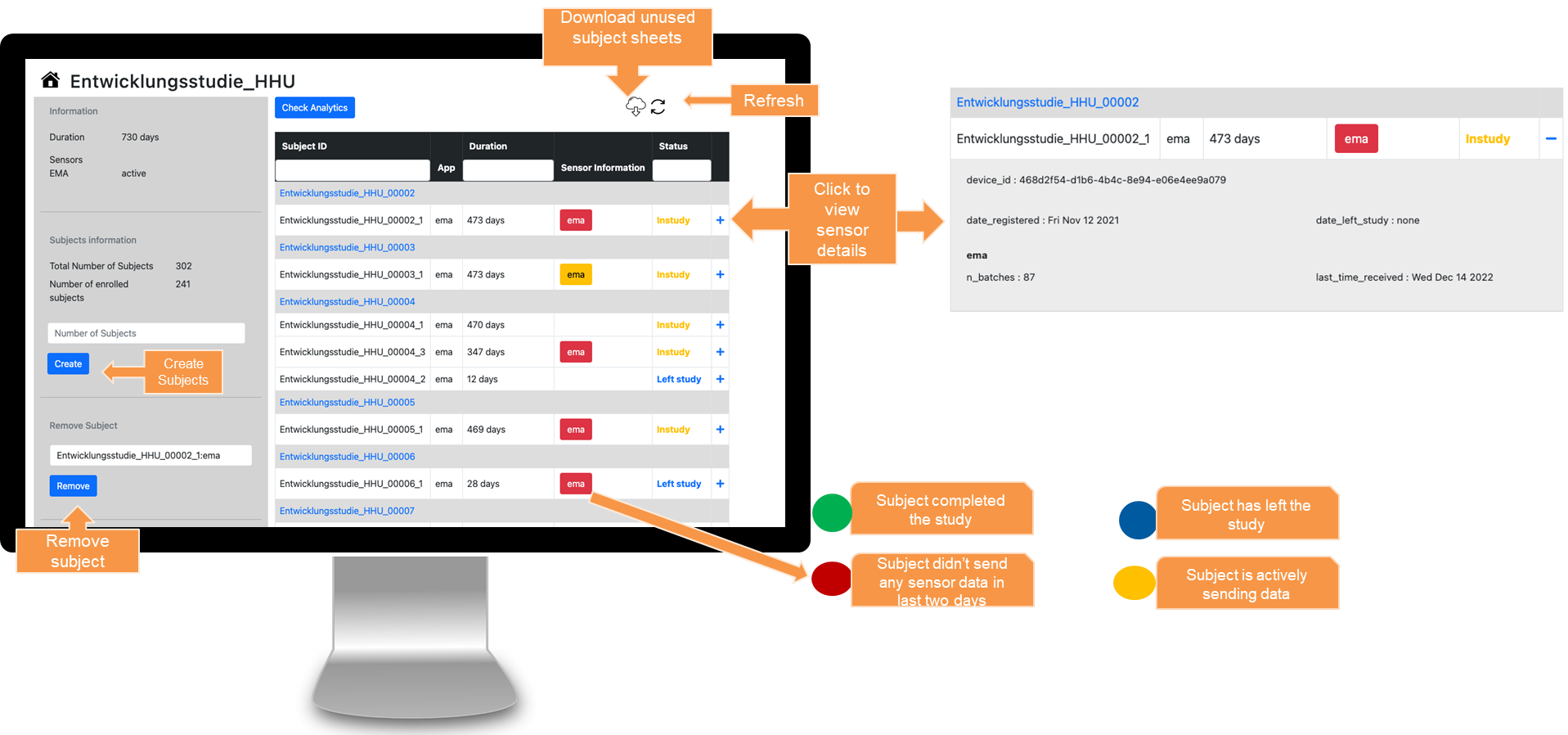
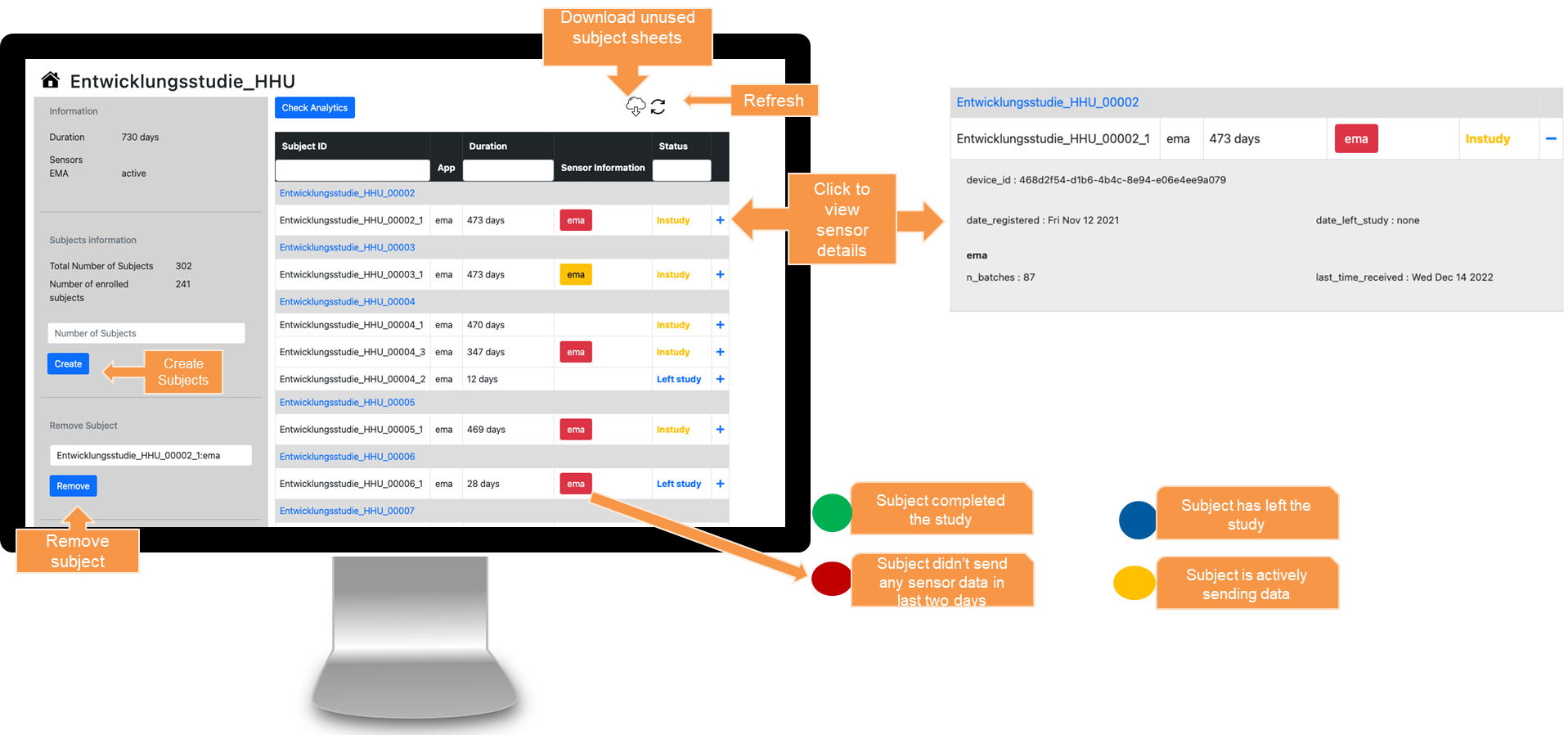
JTrack components: JDash
-
Dashboard for Study Administration
- Investigator's
study & user
management - Data Quality
Control - Notification
Center
|
|
|
Behind the scenes
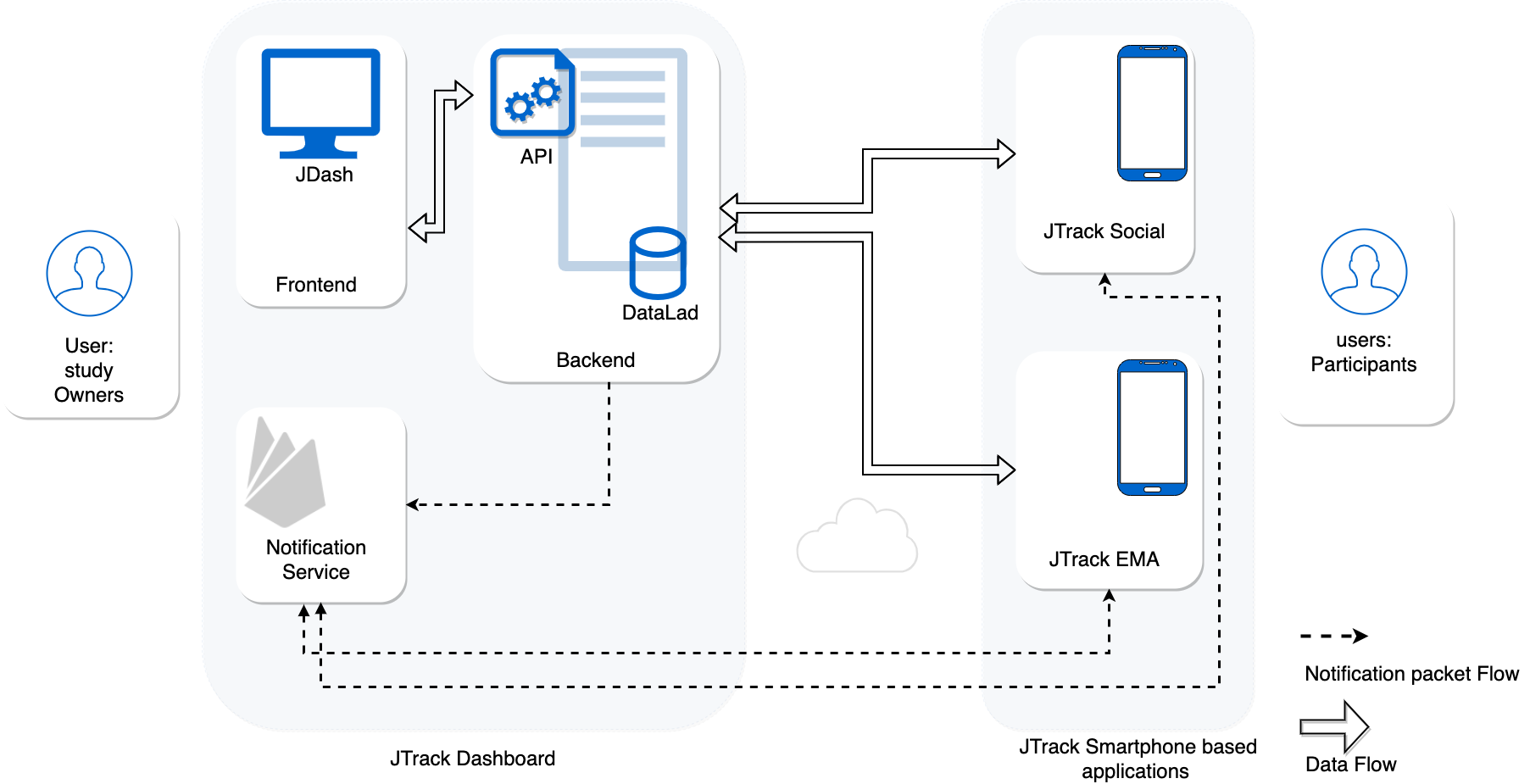
- Servers in Germany
- Data versioning via DataLad
- Authenticated data access via JDash
- Data transfer via HTTPS
Participant's / Investigator's point of view

|

|
Advantages
- Easy to deploy and free environment for collection of real world data (RWD) basically at no cost
- Standardized data collection across centers
- High-density longitudinal data with fully customizable data collection
- Opportunity for citizen science
Acknowledgements
Contact: |
Team:

|

- Open source Python library for easy-to use ML-pipelines, built upon scikit-learn
- Domain-general, but aims to simplify entry into ML for domain scientists with built-in guarantees against most common pitfalls:
- Data leakage
- Overfitting of hyperparameters
- Simplifies common use cases
for supervised ML pipelines,
with feature such as: - Automatic usage of nested cross-
validation for proper evaluation in hyperparameter tuning - Preprocessing based on feature
types, incl. confound removal - Built-in visualization for model
inspection and comparison - Plug-and-play with scikit-learn transformers
| The problem: Expensive AI mistakes |
A solution: User-friendly solutions to common complex use cases |
|
|
Visualization
Interactive "Scores Viewer" for easier model comparison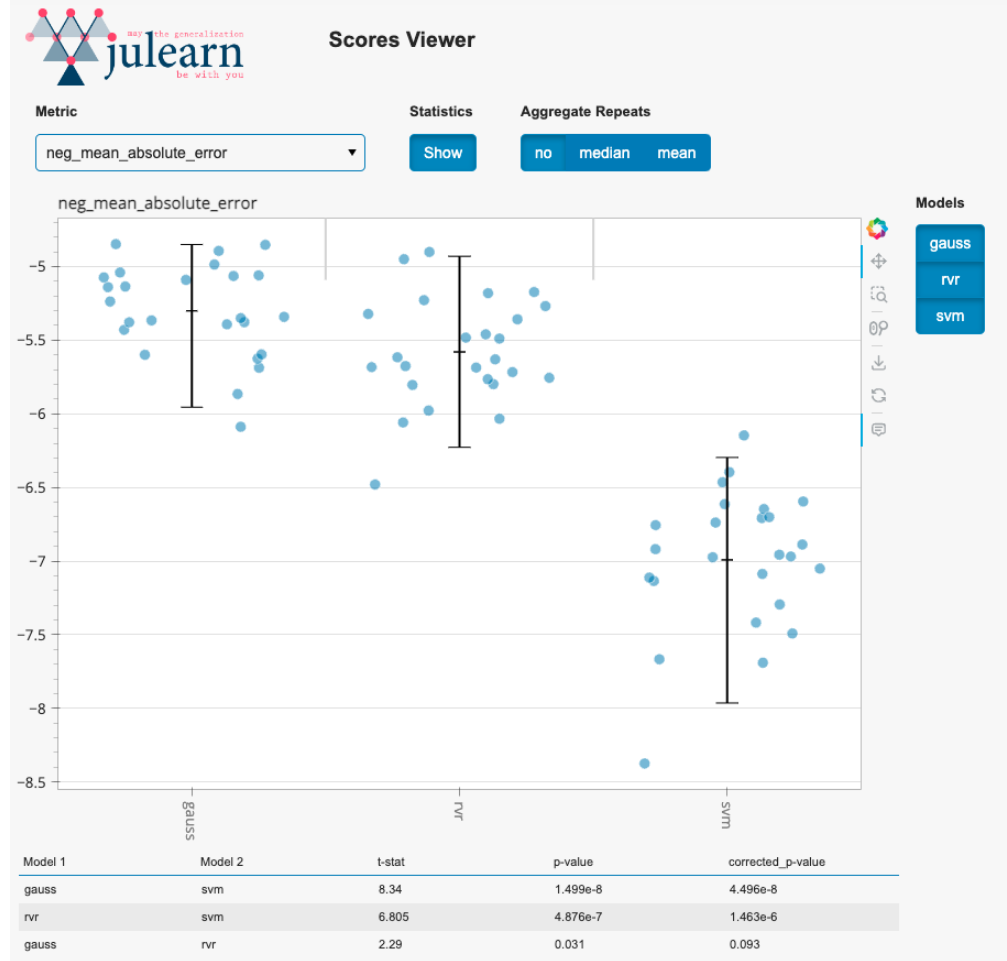
Julearn vs scikit-learn
Simple CV pipelinefrom julearn import run_cross_validation
run_cross_validation(
X=X, y=y, data=data,
preprocess=["zscore"], model="svm",
problem_type="classification",
X_types={"continuous": X} # X_types optional herefrom sklearn.model_selection import cross_validate
from sklearn.svm import SVC # SVR in case of regression
from sklearn.preprocessing import StandardScaler
from sklearn.pipelines import make_pipeline
pipeline = make_pipeline(StandardScaler(), SVC())
cross_validate(X=data.loc[:,X], y=data.loc[:,y], estimator=pipeline)Julearn vs scikit-learn
Nested CV with hyperparameter tuningfrom julearn import run_cross_validation, PipelineCreator
creator=PipelineCreator(problem_type="classification")
creator.add("zscore", with_mean=[True, False])
creator.add("pca", n_components=2)
creator.add("svm", C=[1,2], degree=[3,4])
# X_types optional
run_cross_validation(
X=X, y=y, data=data, model=creator, X_types={"continuous": X})from sklearn.model_selection import cross_validate, GridSearchCV
from sklearn.svm import SVC # SVR in case of regression
from sklearn.preprocessing import StandardScaler
from sklearn.decomposition import PCA
from sklearn.pipelines import make_pipeline
pipeline = make_pipeline(StandardScaler(), PCA(), SVC())
param_grid = {
"standardscaler__with_mean": [True, False],
"pca__n_components": [2],
"scv__C": [1,2],
"svc__degree": [3, 4]
}
grid_pipeline = GridSearchCV(estimator=pipeline, param_grid=param_grid)
cross_validate(X=data.loc[:,X], y=data.loc[:,y], estimator=pipeline)Acknowledgements
- Preprint: Hamdan et al., 2023
- Documentation: juaml.github.io/julearn
- Source Code: github.com/juaml/julearn
 Fede Raimondo
Fede Raimondo
|
 Sami Hamdan
Sami Hamdan
|
 Kaustubh Patil
Kaustubh Patil
|
 Shammi More
Shammi More
|
 Vera Komeyer
Vera Komeyer
|
 Synchon Mandal
Synchon Mandal
|
 Leonard Sasse
Leonard Sasse
|
ML Hours (open to everyone)
Consultancy on Machine-Learning, every second Thursday, 2-4pm
Chat: https://matrix.to/#/#ml:inm7.de
Consultancy on Machine-Learning, every second Thursday, 2-4pm
Chat: https://matrix.to/#/#ml:inm7.de
Even better together
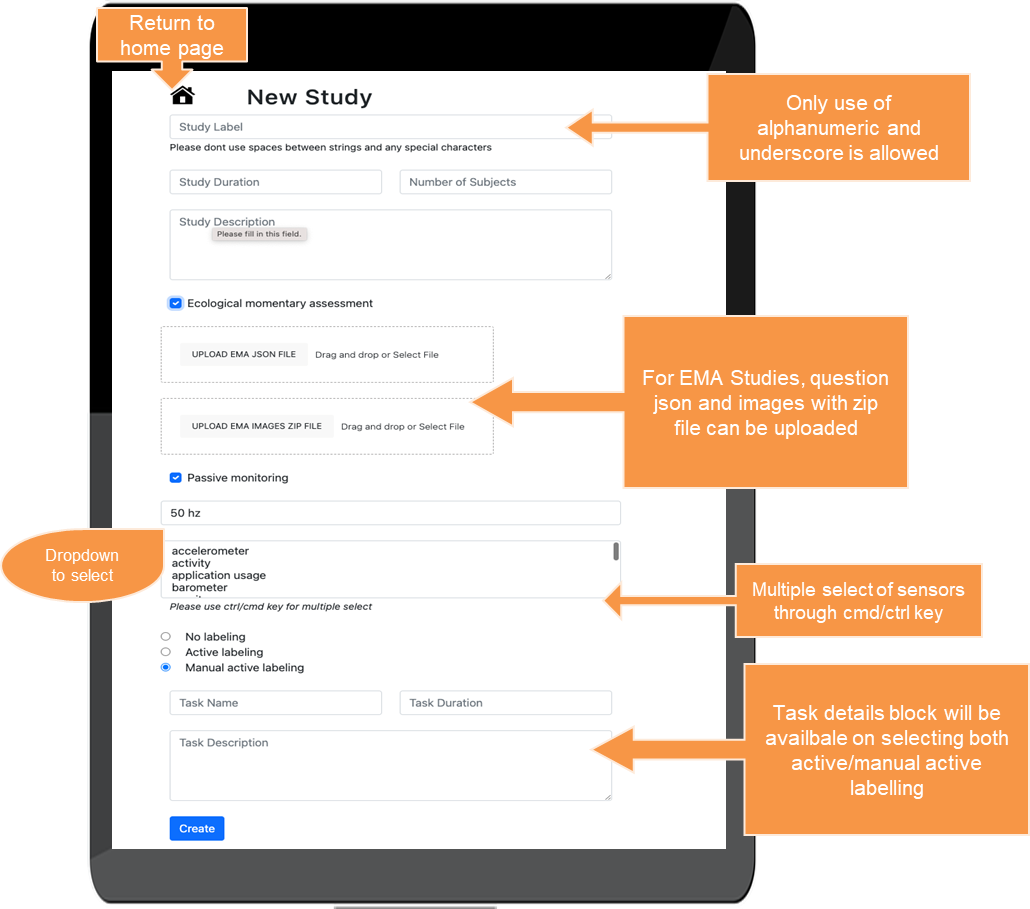
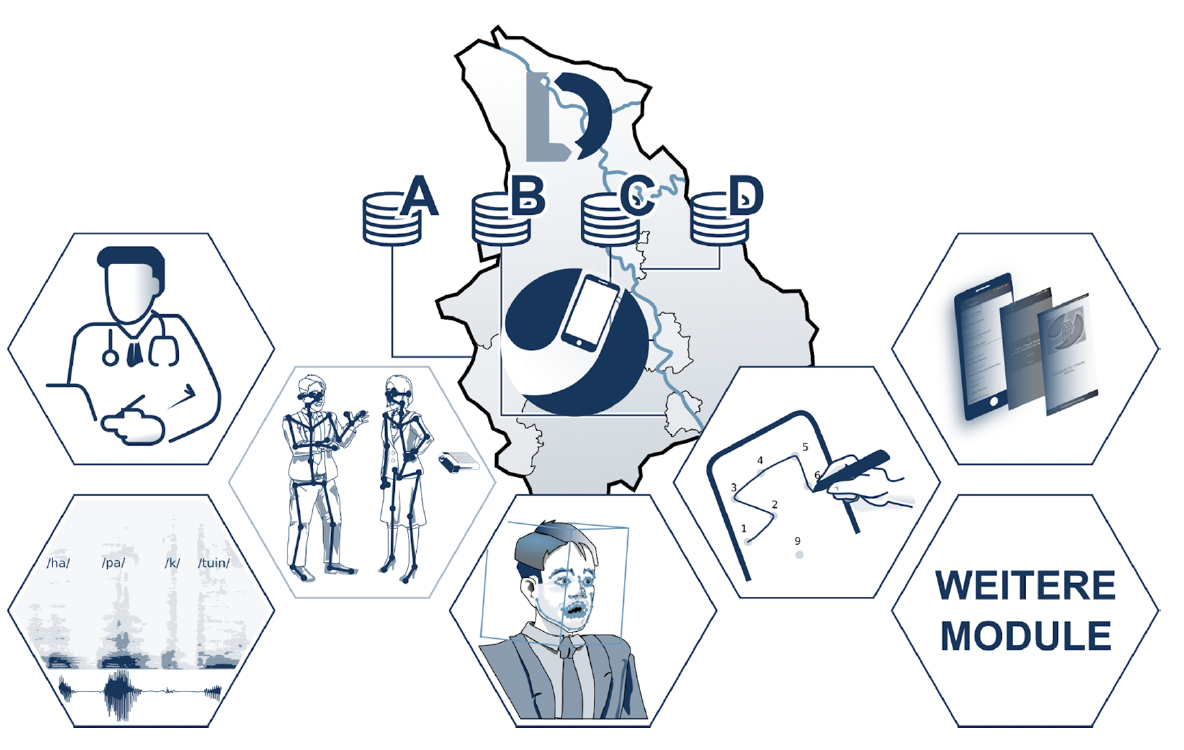
The ABCD-J platform
An open source platform for digital biomarker for neuro-medicine in NRW
-
A collaboration between clinical, academic, and industry partners:
|
Research Center Jülich RWTH Aachen University Bonn University Cologne HHU Düsseldorf LVR Clinics DZNE PeakProfiling CanControl IXP (open to future additions) |
|
Goals
- Social: Promote and facilitate collaboration between multiple centers
- Technical: Accelarate research through homogenization of workflows and processes, with emphasis on digital biomarker development; Elevate existing open technical solutions for research practice adoption
Open research infrastructure
| Clinicians' point of view | Patients' point of view |
|
|
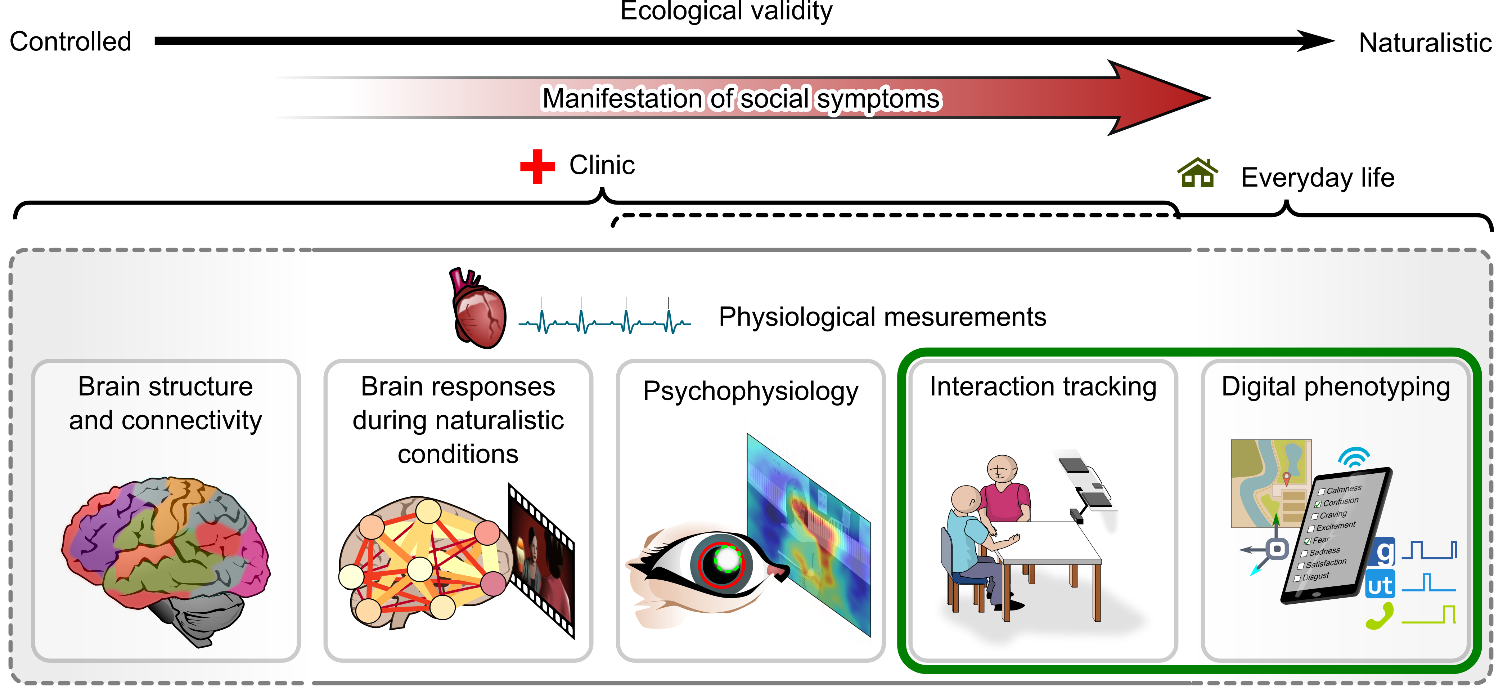
|
|
Open Research Infrastructure
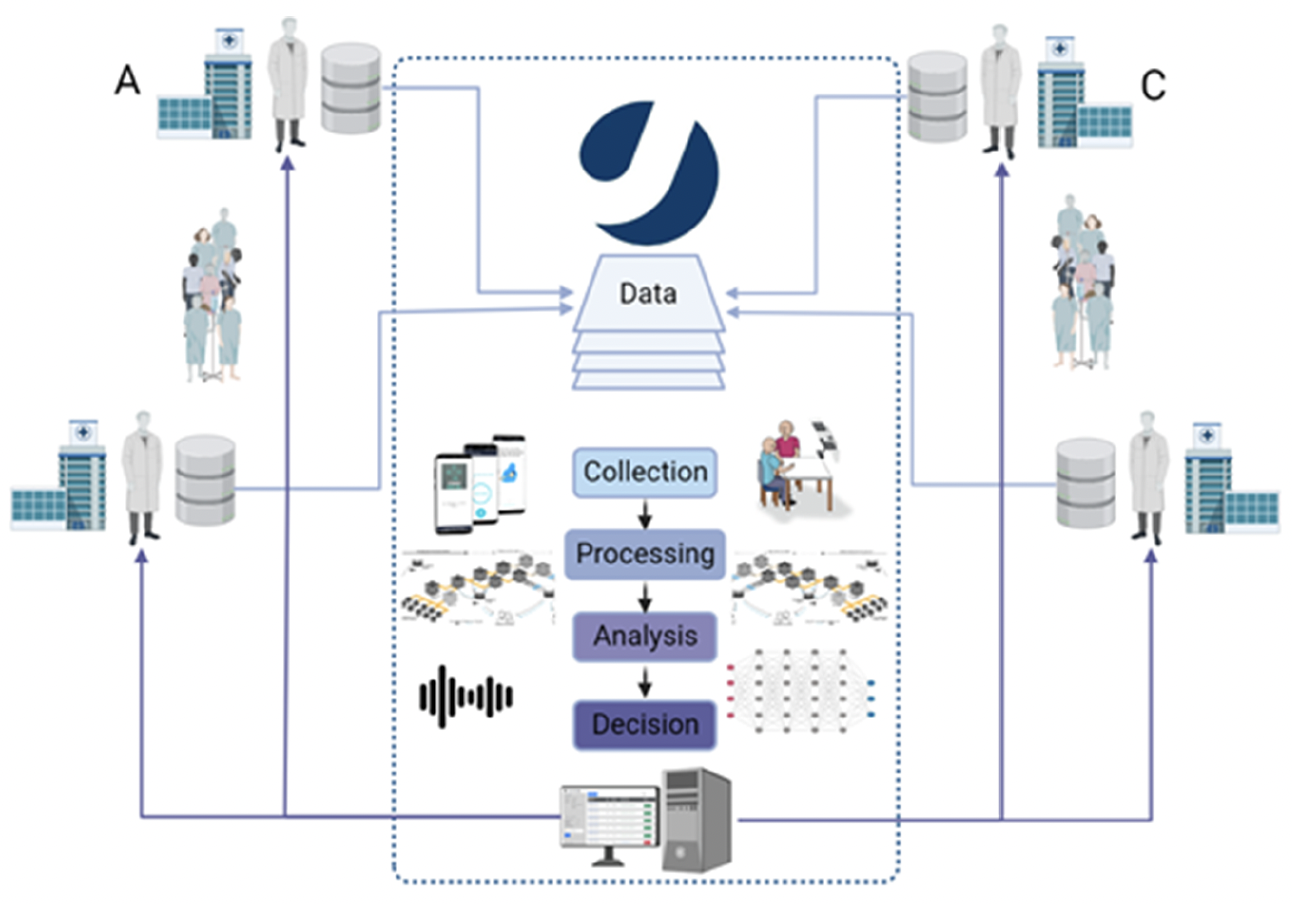
| Research data management's point of view | |
|
|
|
|
|
Front-end and Back-end
- JTrack for decentral, ecologically valid acquisitions, complementing in-clinic assessments
- JDash for study management, participant management, and analytics overview (derived study data at subject & group level)
- Central data overview and analytics at FZJ
- Provenance-tracked analysis and modeling
- Automated meta-data extraction for data discoverability
- Result overview for clinical decision making
|
|
Opportunities
Software improves with its use cases- JTrack integration into different types wearables
- JTrack integration of cognitive tasks and feedback to participants
- Julearn integration into JDash
- More meta-data extractors for DataLad
- ...
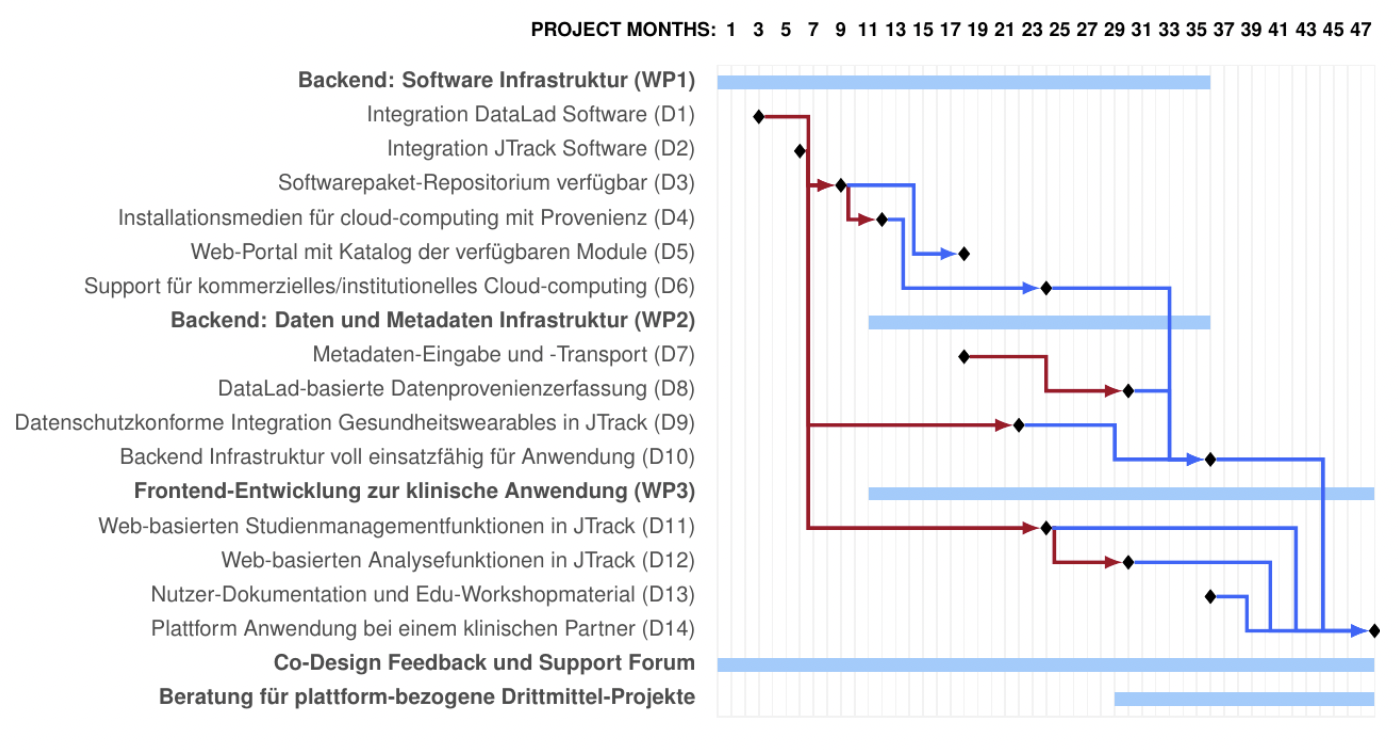
Current (first) steps
-
Data cataloging
- Leveraging legacy data via data census and meta-data catalog
- improved discovery without direct data transfer
- homogenization of access request procedures
- establishing a legal basis for (re-)use
- Example: data.sfb1451.de
- Demonstrator for data infrastructure based on §21 data (standardized and anonymized performance data of hospitals, legally required, submitted yearly to InEK by all hospitals)
- Recommendations for common digital tools and workflows for common tasks and processes
- Selection of digital measures from clinical routine for first trials
Feasibility/Proof-of-concept study
Summary
- Open Source (Research) Software aids in various domain-general or -specific applications.
- Open Science needs Open Source: For transparency and reproducibility, for science-specific requirements, for open formats, for re-use, and to enable interoperability across tools.
- Collaboration across clinical and research settings is a technical, social, and legal challenge. Technical solutions won't save us alone, but they are a good first step.
- We build clinical research infrastructure on open tools, for better science
Thanks!
Questions?
Inputs, Resources, Synergies