DataLad
Decentralized Management of Digital Objects for Open Science
| Dr. Adina Wagner
|
|
|
Institute of Neuroscience and Medicine, Brain & Behavior (INM-7) Research Center Jülich |
 |
Slides: DOI 10.5281/zenodo.15193934 (Scan the QR code)
files.inm7.de/adina/talks/html/nhr_2025_datalad.html
Acknowledgements
|
Funders







Collaborators







|
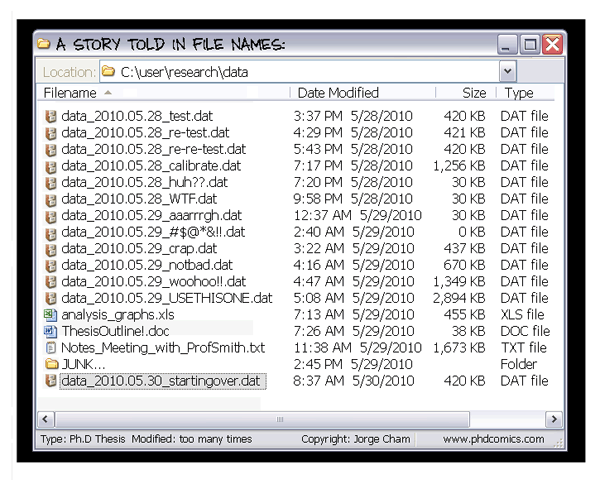
The building blocks of a scientific result are rarely static
Data changes
(errors are fixed, data is extended,
naming standards change, an analysis
requires only a subset of your data...)
- Domain-agnostic command-line tool (+ graphical user interface), built on top of Git & Git-annex
- Open source (MIT) research software developed since 2013
- Available for all major operating systems
- Major features:
- Version-controlling arbitrarily large content
- Version control data & software alongside to code!
- Transport mechanisms for sharing & obtaining data
- Consume & collaborate on data (analyses) like software
- (Computationally) reproducible data analysis
- Track and share provenance of all digital objects
- (... and much more)

A DataLad dataset is a joint Git/git-annex repository that can version control any file
|
|

Which data (at which version), with which code, running with what parameterization in which computational environment, to generate what?
|
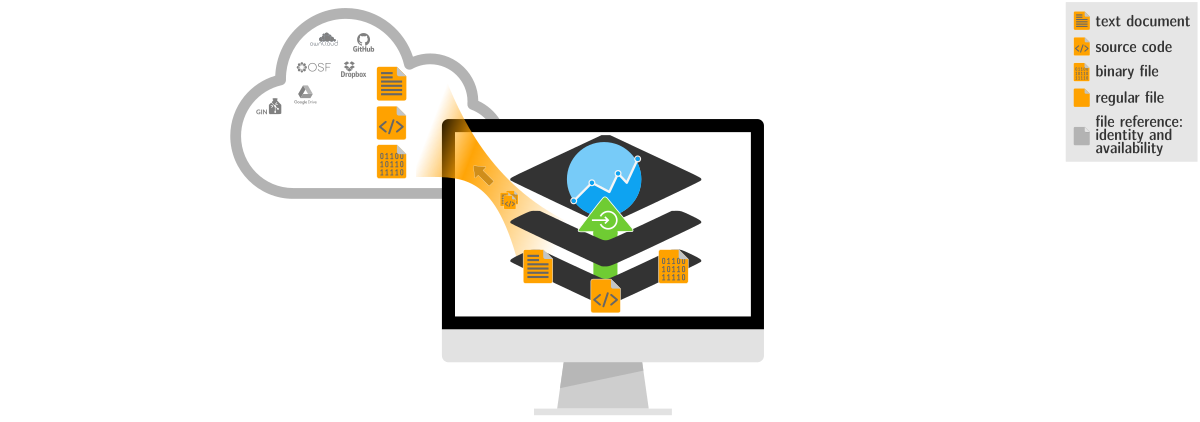
Decentral data transport to Git hosting, local or remote infrastructure, or external hosting services
|

Outcomes can be validated. This enables audits, promotes accountability, and streamlines automated "upgrades" of outputs
|
|

Datasets can be (re-)used as modular components in larger contexts — propagating their traits. They are verifiable, portable, self-contained data structures
|
Version control beyond text files
 Using git-annex,
DataLad version controls large data
Using git-annex,
DataLad version controls large data
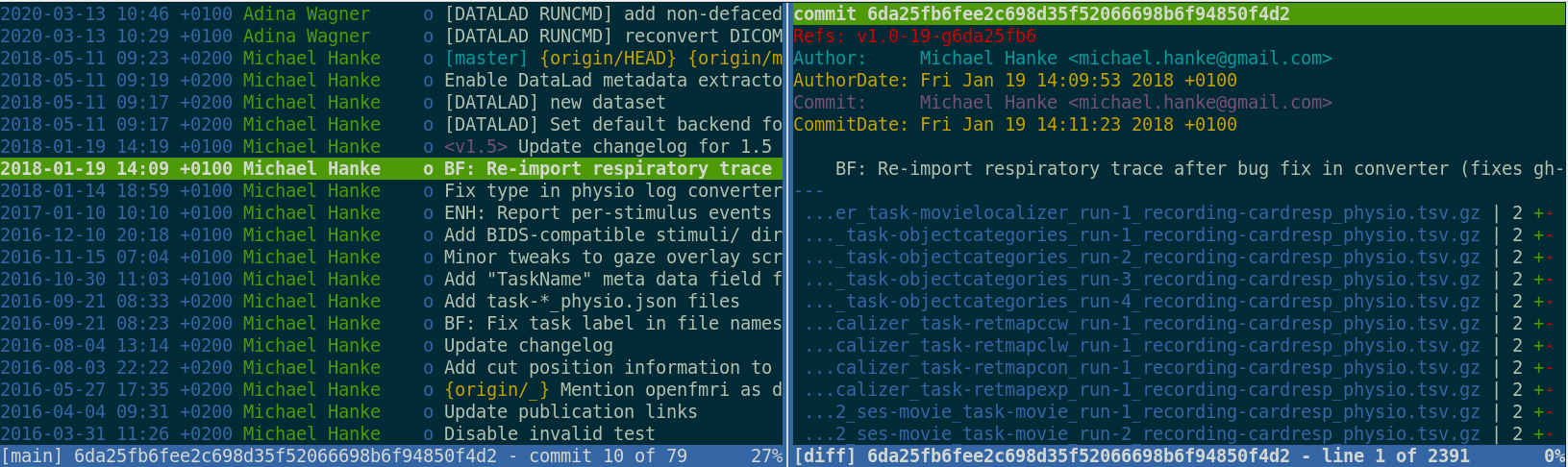
Version control beyond text files
- Datasets have an annex to track files without placing their content into Git
- Rather than content, identity (hash) and location information is put into Git:
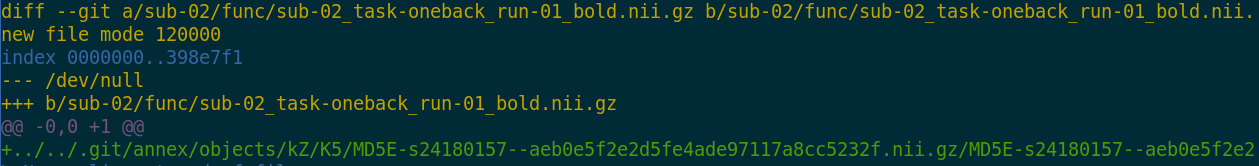
- Where the filesystem allows it, annexed files are symlinks:
$ ls -l sub-02/func/sub-02_task-oneback_run-01_bold.nii.gz
lrwxrwxrwx 1 adina adina 142 Jul 22 19:45 sub-02/func/sub-02_task-oneback_run-01_bold.nii.gz ->
../../.git/annex/objects/kZ/K5/MD5E-s24180157--aeb0e5f2e2d5fe4ade97117a8cc5232f.nii.gz/MD5E-s24180157
--aeb0e5f2e2d5fe4ade97117a8cc5232f.nii.gz
- The symlink reveals: This internal data organization based on identity hash
$ md5sum sub-02/func/sub-02_task-oneback_run-01_bold.nii.gz
aeb0e5f2e2d5fe4ade97117a8cc5232f sub-02/func/sub-02_task-oneback_run-01_bold.nii.gz
- The (tiny) symlink instead of the (potentially large) file content is
committed - version controlling precise file identity without checking contents into Git
- File availability information is stored to record a decentral network of file content. A file can exist in multiple different locations.
$ git annex whereis sub-02/func/sub-02_task-oneback_run-01_bold.nii.gz
whereis sub-02/func/sub-02_task-oneback_run-01_bold.nii.gz (2 copies)
8c3680dd-6165-4749-adaa-c742232bc317 -- git@8242caf9acd8:/data/repos/adswa/bidsdata.git [gin]
fff8fdbc-3185-4b78-bd12-718717588442 -- adina@muninn:~/bids-data [here]
ok
Git versus Git-annex
- Data in datasets is either stored in Git or git-annex
- By default, everything is annexed, i.e., stored in a dataset annex
| Git | git-annex |
| handles small files well (text, code) | handles all types and sizes of files well |
| file contents are in the Git history and will be shared upon git/datalad push | file contents are in the annex. Not necessarily shared |
| Shared with every dataset clone | Can be kept private on a per-file level when sharing the dataset |
| Useful: Small, non-binary, frequently modified, need-to-be-accessible (DUA, README) files | Useful: Large files, private files |
(Raw) data mismanagement
- Multiple large datasets are available on a compute cluster 🏞
- Each researcher creates their own copies of data ⛰
- Multiple different derivatives and results are computed from it 🏔
- Data, copies of data, half-baked data transformations, results, and old versions of results are kept - undocumented 🌋
Share data like source code
Transport logistics: Lots of data, little disk-usage
- Cloned datasets are lean. "Meta data" (file names, availability) are present, but no file content:
$ datalad clone git@github.com:psychoinformatics-de/studyforrest-data-phase2.git
install(ok): /tmp/studyforrest-data-phase2 (dataset)
$ cd studyforrest-data-phase2 && du -sh
18M .$ datalad get sub-01/ses-movie/func/sub-01_ses-movie_task-movie_run-1_bold.nii.gz
get(ok): /tmp/studyforrest-data-phase2/sub-01/ses-movie/func/
sub-01_ses-movie_task-movie_run-1_bold.nii.gz (file) [from mddatasrc...]- Have access to more data on your computer than you have disk-space:
# eNKI dataset (1.5TB, 34k files):
$ du -sh
1.5G .
# HCP dataset (~200TB, >15 million files)
$ du -sh
48G . Publishing datasets
-
Publish datasets, their annexed contents, or both to infrastructure of your choice
Interoperability
- DataLad is built to maximize interoperability and streamline routines across hosting and storage technology
Dataset Nesting
- Seamless nesting mechanisms:
- hierarchies of datasets in super-/sub-dataset relationships
- based on Git submodules, but more seamless: Mono-repo feel thanks to recursive operations
- Overcomes scaling issues with large amounts of files
adina@bulk1 in /ds/hcp/super on git:master❱ datalad status --annex -r
15530572 annex'd files (77.9 TB recorded total size)
nothing to save, working tree cleanIntuitive data analysis structure
$ cd myanalysis
# we can install analysis input data as a subdataset to the dataset
$ datalad clone -d . https://github.com/datalad-handbook/iris_data.git input/
[INFO ] Scanning for unlocked files (this may take some time)
[INFO ] Remote origin not usable by git-annex; setting annex-ignore
install(ok): input (dataset)
add(ok): input (file)
add(ok): .gitmodules (file)
save(ok): . (dataset)
action summary:
add (ok: 2)
install (ok: 1)
save (ok: 1)
Leaving a trace
"Shit, which version of which script produced these outputs from which version of what data?"
"Shit, why buttons did I click and in which order did I use all those tools?"

Leaving a trace
datalad run wraps around anything expressed in a command line call and saves the dataset modifications resulting from the execution.
datalad rerun repeats captured executions. If the outcomes differ, it saves a new state of them.
datalad containers-run executes command line calls inside a tracked software container and saves the dataset modifications resulting from the execution.
data analysis provenance
Enshrine the analysis in a script
Here: extract_lc_timeseries.py
$ datalad containers-run \
--message "Time series extraction from Locus Coeruleus"
--container-name nilearn \
--input 'mri/*_bold.nii' \
--output 'sub-*/LC_timeseries_run-*.csv' \
"python3 code/extract_lc_timeseries.py"
-- Git commit --
commit 5a7565a640ff6de67e07292a26bf272f1ee4b00e
Author: Adina Wagner adina.wagner@t-online.de
AuthorDate: Mon Nov 11 16:15:08 2019 +0100
Commit: Adina Wagner adina.wagner@t-online.de
CommitDate: Mon Nov 11 16:15:08 2019 +0100
[DATALAD RUNCMD] Time series extraction from Locus Coeruleus
=== Do not change lines below ===
{
"cmd": "singularity exec --bind {pwd} .datalad/environments/nilearn.simg bash..",
"dsid": "92ea1faa-632a-11e8-af29-a0369f7c647e",
"inputs": [
"mri/*.bold.nii.gz",
".datalad/environments/nilearn.simg"
],
"outputs": ["sub-*/LC_timeseries_run-*.csv"],
...
}
^^^ Do not change lines above ^^^
---
sub-01/LC_timeseries_run-1.csv | 1 +
...
data analysis provenance
Record code execution together
with
input-data, output files and software
environment in the
execution-command
$ datalad containers-run \
--message "Time series extraction from Locus Coeruleus"
--container-name nilearn \
--input 'mri/*_bold.nii' \
--output 'sub-*/LC_timeseries_run-*.csv' \
"python3 code/extract_lc_timeseries.py"
-- Git commit --
commit 5a7565a640ff6de67e07292a26bf272f1ee4b00e
Author: Adina Wagner adina.wagner@t-online.de
AuthorDate: Mon Nov 11 16:15:08 2019 +0100
Commit: Adina Wagner adina.wagner@t-online.de
CommitDate: Mon Nov 11 16:15:08 2019 +0100
[DATALAD RUNCMD] Time series extraction from Locus Coeruleus
=== Do not change lines below ===
{
"cmd": "singularity exec --bind {pwd} .datalad/environments/nilearn.simg bash..",
"dsid": "92ea1faa-632a-11e8-af29-a0369f7c647e",
"inputs": [
"mri/*.bold.nii.gz",
".datalad/environments/nilearn.simg"
],
"outputs": ["sub-*/LC_timeseries_run-*.csv"],
...
}
^^^ Do not change lines above ^^^
---
sub-01/LC_timeseries_run-1.csv | 1 +
...
data analysis provenance
Result: machine readable record about which data, code, and
software produced a result how, when, and why.
$ datalad containers-run \
--message "Time series extraction from Locus Coeruleus"
--container-name nilearn \
--input 'mri/*_bold.nii' \
--output 'sub-*/LC_timeseries_run-*.csv' \
"python3 code/extract_lc_timeseries.py"
-- Git commit --
commit 5a7565a640ff6de67e07292a26bf272f1ee4b00e
Author: Adina Wagner adina.wagner@t-online.de
AuthorDate: Mon Nov 11 16:15:08 2019 +0100
Commit: Adina Wagner adina.wagner@t-online.de
CommitDate: Mon Nov 11 16:15:08 2019 +0100
[DATALAD RUNCMD] Time series extraction from Locus Coeruleus
=== Do not change lines below ===
{
"cmd": "singularity exec --bind {pwd} .datalad/environments/nilearn.simg bash..",
"dsid": "92ea1faa-632a-11e8-af29-a0369f7c647e",
"inputs": [
"mri/*.bold.nii.gz",
".datalad/environments/nilearn.simg"
],
"outputs": ["sub-*/LC_timeseries_run-*.csv"],
...
}
^^^ Do not change lines above ^^^
---
sub-01/LC_timeseries_run-1.csv | 1 +
...
data analysis provenance
Use the unique identifier of the execution record
$ datalad rerun 5a7565a640ff6de67
[INFO ] run commit 5a7565a640ff6de67; (Time series extraction from Locus Coeruleus)
[INFO ] Making sure inputs are available (this may take some time)
get(ok): mri/sub-01_bold.nii (file)
get(ok): mri/sub-02_bold.nii (file)
[...]
[INFO ] == Command start (output follows) =====
[INFO ] == Command exit (modification check follows) =====
add(ok): sub-01/LC_timeseries_run-*.csv(file)
add(ok): sub-02/LC_timeseries_run-*.csv (file)
[...]
action summary:
add (ok: 30)
get (ok: 30)
save (ok: 2)
unlock (ok: 30)
data analysis provenance
... to have a machine recompute and verify past work
$ datalad rerun 5a7565a640ff6de67
[INFO ] run commit 5a7565a640ff6de67; (Time series extraction from Locus Coeruleus)
[INFO ] Making sure inputs are available (this may take some time)
get(ok): mri/sub-01_bold.nii (file)
get(ok): mri/sub-02_bold.nii (file)
[...]
[INFO ] == Command start (output follows) =====
[INFO ] == Command exit (modification check follows) =====
add(ok): sub-01/LC_timeseries_run-*.csv(file)
add(ok): sub-02/LC_timeseries_run-*.csv (file)
[...]
action summary:
add (ok: 30)
get (ok: 30)
save (ok: 2)
unlock (ok: 30)
DataLad for scientific workflows?
- Scientific building blocks are not static.
- Version control beyond text
- Science is build from modular units.
- Nesting
- Science is exploratory, iterative, multi-stepped, and complex.
- Provenance
- Science is collaborative.
- Transport logistics
Research data management is tied to reproducibility

FAIRly big setup
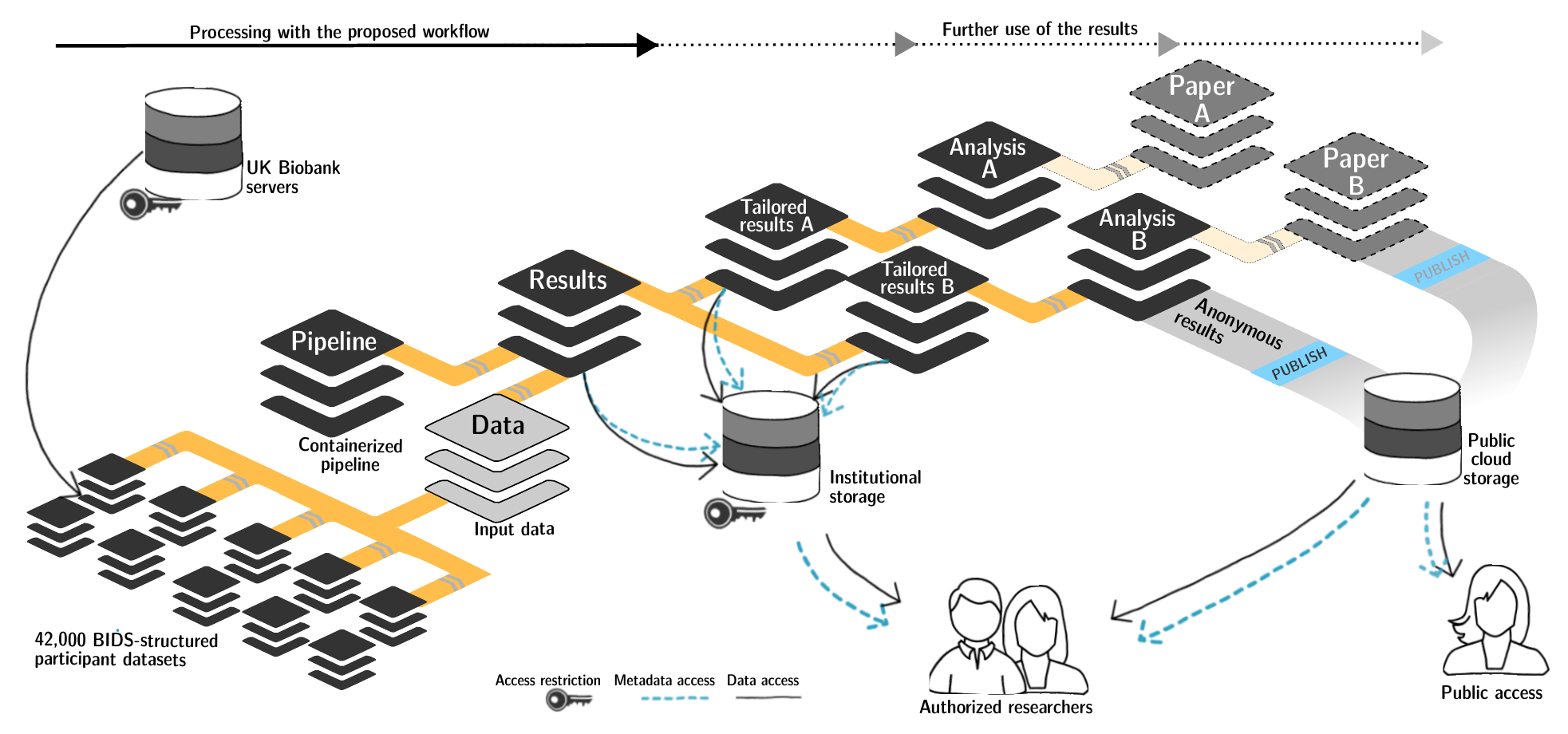
-
Exhaustive tracking
- datalad-ukbiobank extension downloads, transforms & track the evolution of the complete data release in DataLad datasets
- Native and BIDSified data layout (at no additional disk space usage)
- Structured in 42k individual datasets, combined to one superdataset
- Containerized pipeline in a software container
- Link input data & computational pipeline as dependencies
Wagner, Waite, Wierzba et al. (2021). FAIRly big: A framework for computationally reproducible processing of large-scale data.
FAIRly big workflow
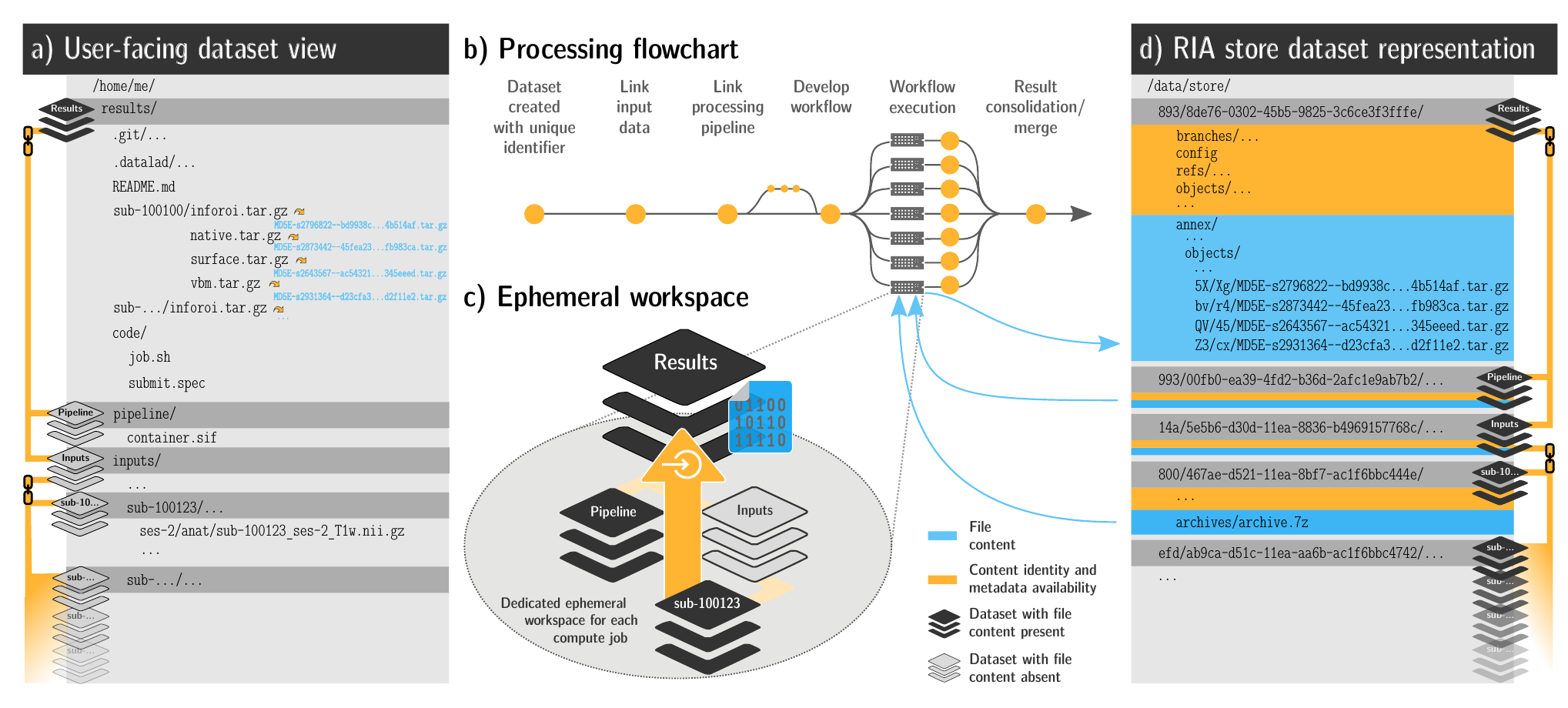
-
portability
- Parallel processing: 1 job = 1 subject (number of concurrent jobs capped at the capacity of the compute cluster)
- Each job is computed in a ephemeral (short-lived) dataset clone, results are pushed back: Ensure exhaustive tracking & portability during computation
- Content-agnostic persistent (encrypted) storage (minimizing storage and inodes)
- Common data representation in secure environments
Wagner, Waite, Wierzba et al. (2021). FAIRly big: A framework for computationally reproducible processing of large-scale data.
FAIRly big provenance capture
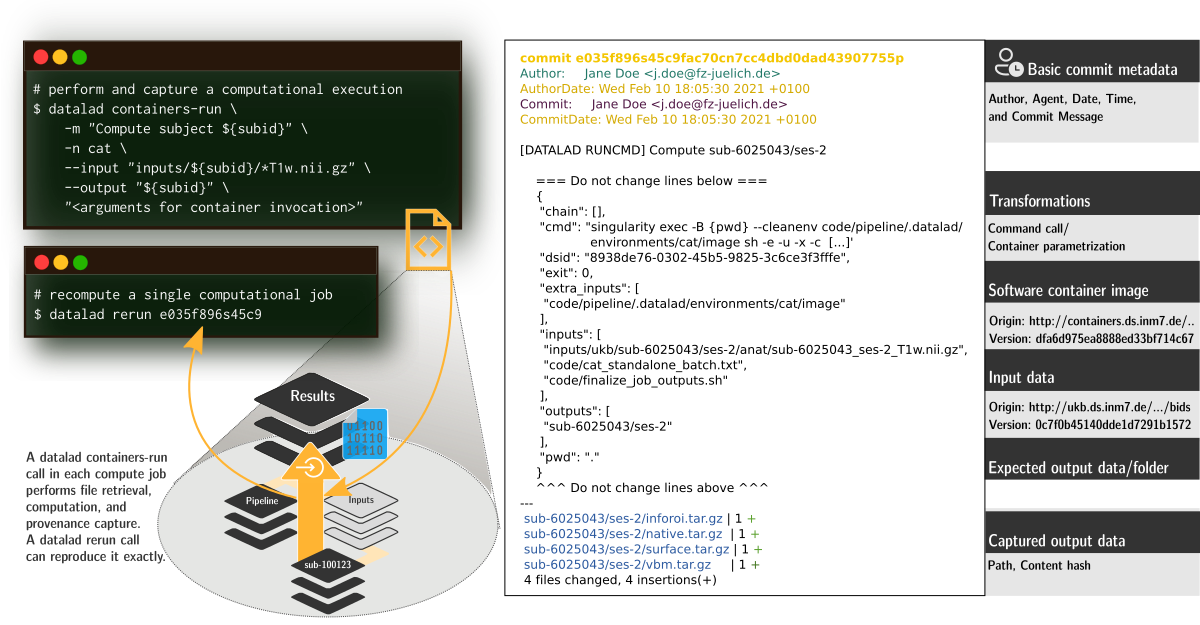
-
Provenance
- Every single pipeline execution is tracked
- Execution in ephemeral workspaces ensures results individually reproducible without HPC access
Wagner, Waite, Wierzba et al. (2021). FAIRly big: A framework for computationally reproducible processing of large-scale data.
Current and future developments
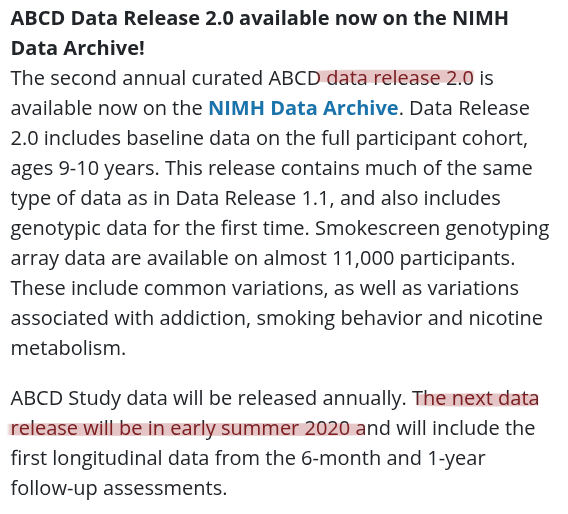
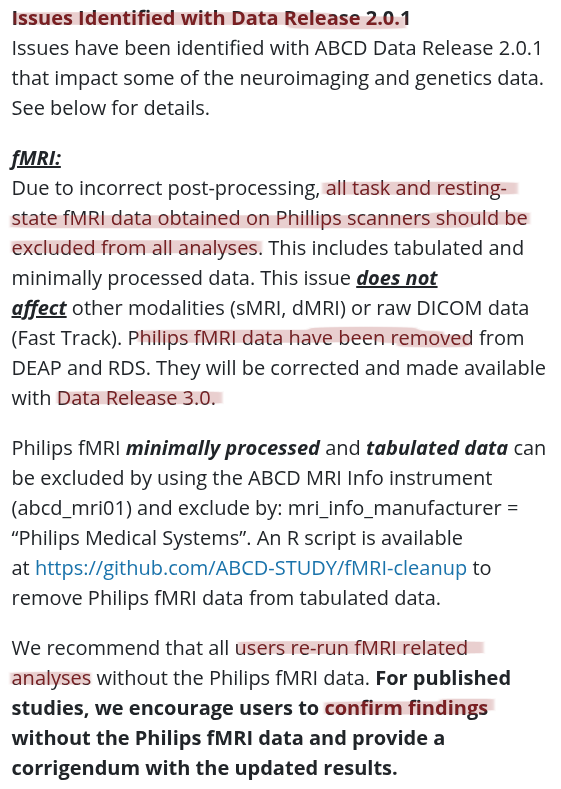
The building blocks of a scientific result are
... sometimes unreliable or threatened
Data changes
Due to presidential executive orders
to remove files mentioning "gender"
Freedom? Chose Decentralization
- Infrastructure is ephemeral:
- Change of institutional contracts
- Change of affiliations
- Geopolitical developments?
- DataLad datasets are portable
- Effortless migrations to different Git or data hosting
- Versioning allows for integrity checks
Delineation and advantages of decentral versus central RDM:
Hanke et al., (2021). In defense of decentralized research data management
Going self-hosted with forgejo-aneksajo
- Forgejo (forgejo.org): Fork of Gitea
- Forgejo-aneksajo: Forgejo with git-annex support

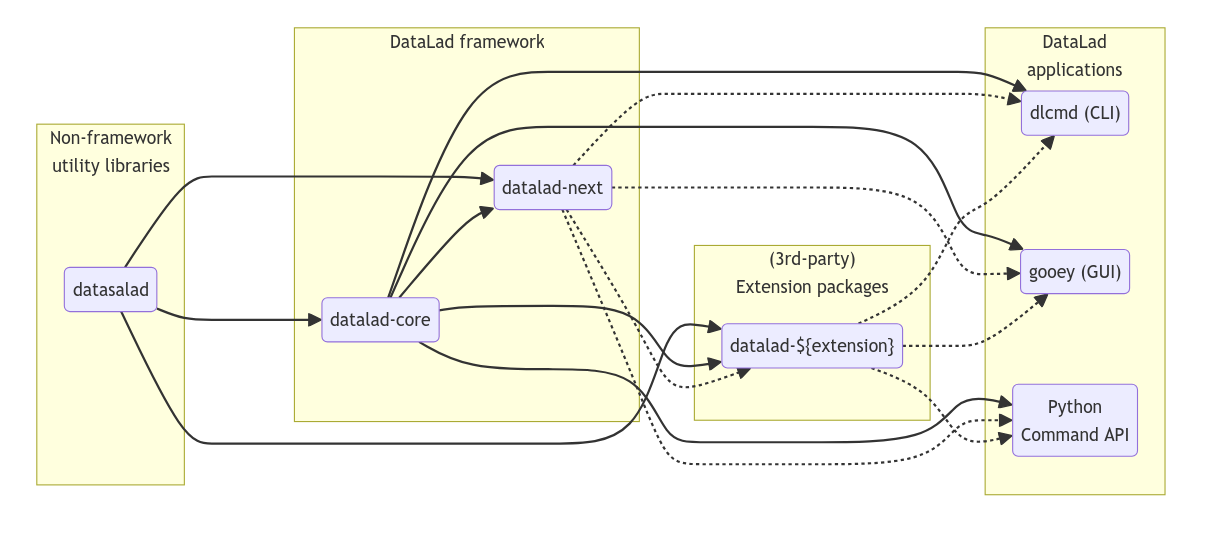
Development Roadmap
Join us!
-
Distribits 2025
- International conference on technologies for distributed data management
- 2 day conference plus single-day Hackathon
- @ Haus der Universität Düsseldorf
- Registration open until May 1st
distribits.live
DataLad contact and more information
| Website + Demos | http://datalad.org |
| Documentation | http://handbook.datalad.org |
| Talks and tutorials | https://youtube.com/datalad |
| Development | http://github.com/datalad |
| Support | https://matrix.to/#/#datalad:matrix.org |
| Open data | http://datasets.datalad.org |
| Mastodon | @datalad@fosstodon.org |
Thanks!