Research Data Management with DataLad
🚀
for easier, open, and transparent science
Adina Wagner
 @AdinaKrik
@AdinaKrik |
|
|
Psychoinformatics lab,
Institute of Neuroscience and Medicine, Brain & Behavior (INM-7) Research Center Jülich |
Research data management (RDM)
- (Research) Data = every digital object involved in your project: code, software/tools, raw data, processed data, results, manuscripts ...
- Data needs to be managed FAIRly- from creation to use, publication, sharing, archiving, re-use, or destruction:

- Research data management is a key component for reproducibility, efficiency, and impact/reach of data analysis projects
Why data management?

⬆
This a metaphor for most projects after publication
Why data management?
This a metaphor for reproducing (your own) research
a few months after publication
⬇

Why data management?
| This is a metaphor for many computational ➡ clusters without RDM |
 |
Why data management? Different view points
- "Oh well if others say so": External requirements and expectations
- Funders & publishers require it
- Scientific peers increasingly expect it
- "There is no other way": Some datasets require it
- Exciting datasets (UKBiobank, HCP, ...) are so large that neither computational infrastructure nor typical analysis workflows scale to their sizes
- "OMG when can I start?": Intrinsic motivation and personal & scientific benefits
- The quality, efficiency and replicability of your work improves
|
|

|
|
|

|
|
|

|
(all of those are valid reasons for RDM, but its fun if you have Minion-attitude)
Today

- General overview of DataLad
- Hands-on experience: Copy-Paste code snippets at handbook.datalad.org/en/latest/code_from_chapters/MPI_code.html
- DataLad-centric solutions to real-life data management problems
Further resources
- Everything I'm talking about is documented in text and video tutorials, and you can reach out for any questions!
- Comprehensive user documentation in the DataLad Handbook (handbook.datalad.org)
- Recordings of talks and tutorials on our YouTube channel
- Reach out with questions via Matrix or GitHub (github/datalad/datalad or github/datalad-handbook/book)
polling system for live-feedback
Let's start
Requirements
- DataLad version 0.12.x or later (Installation instructions at handbook.datalad.org)
- A configured Git identity:
$ git config --add user.name "Bob McBobface"
$ git config --add user.email bob@example.com - (You have about 5 minutes to still install it)
Acknowledgements
|
Funders






Collaborators





|
 Core Features:
Core Features:
- Joint version control (Git, git-annex) for code, software, and data
- Provenance capture: Create and share machine-readable, re-executable records of your data analysis for reproducible, transparent, and FAIR research
- Data transport mechanisms: Install or share complete projects extremely lightweight, retrieve data on demand and drop it to free up space without losing data access or provenance, collaborate remotely on scientific projects
Examples of what DataLad can be used for:
- Publish or consume datasets via GitHub, GitLab, OSF, or similar services
Examples of what DataLad can be used for:
- Creating and sharing reproducible, open science: Sharing data, software, code, and provenance
Examples of what DataLad can be used for:
- Behind-the-scenes infrastructure component for data transport and versioning (e.g., used by OpenNeuro, brainlife.io , the Canadian Open Neuroscience Platform (CONP), CBRAIN)
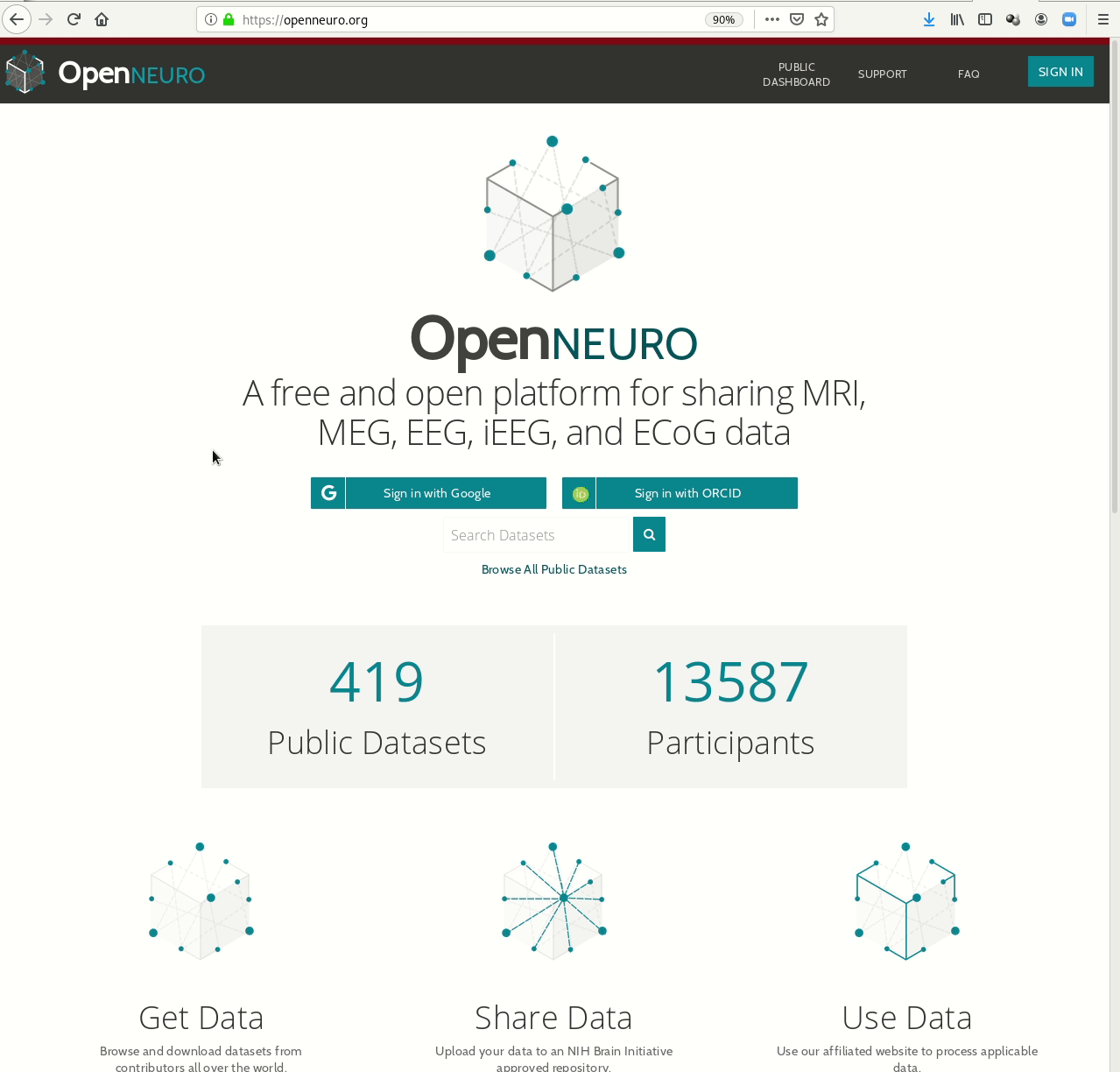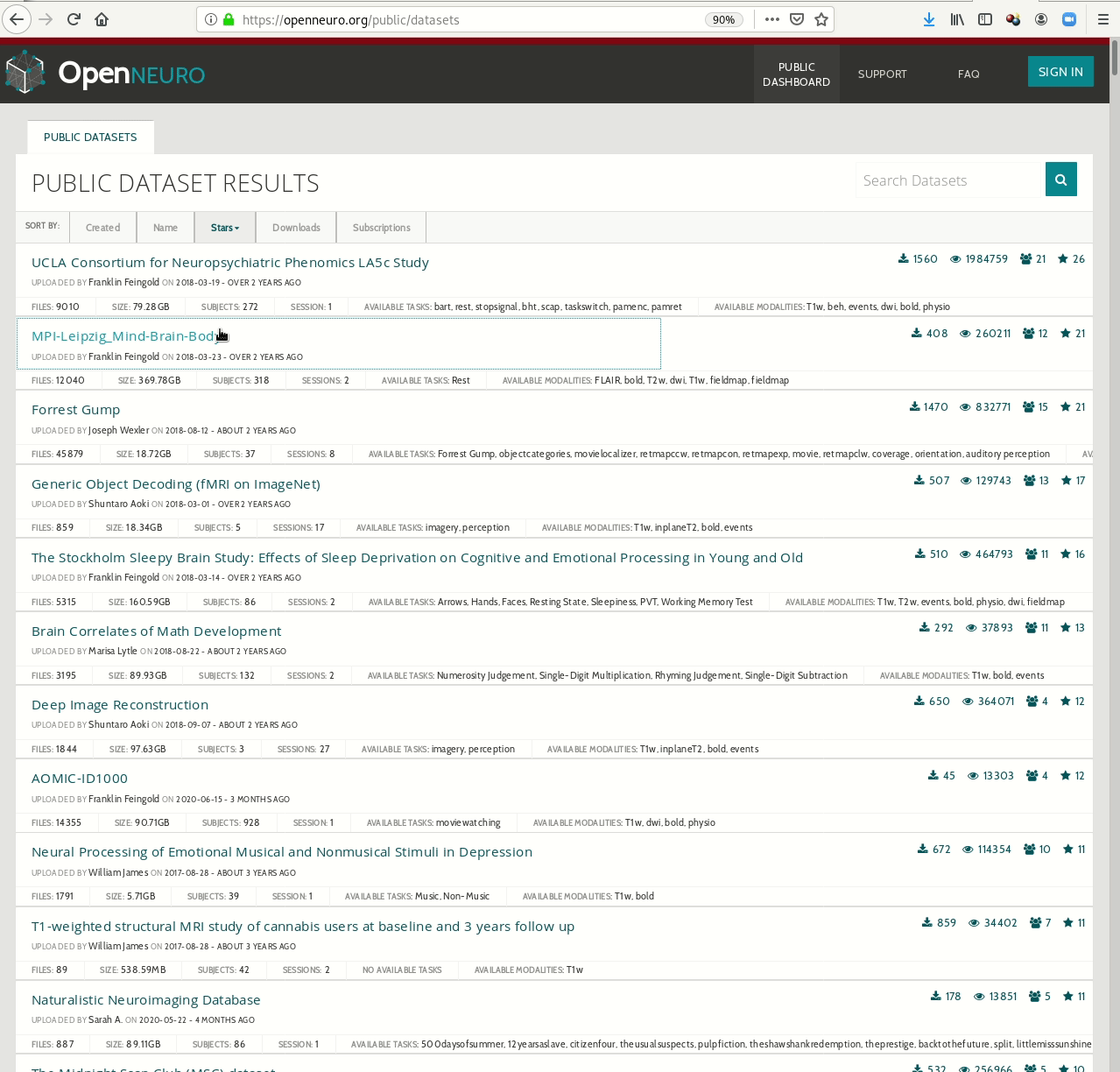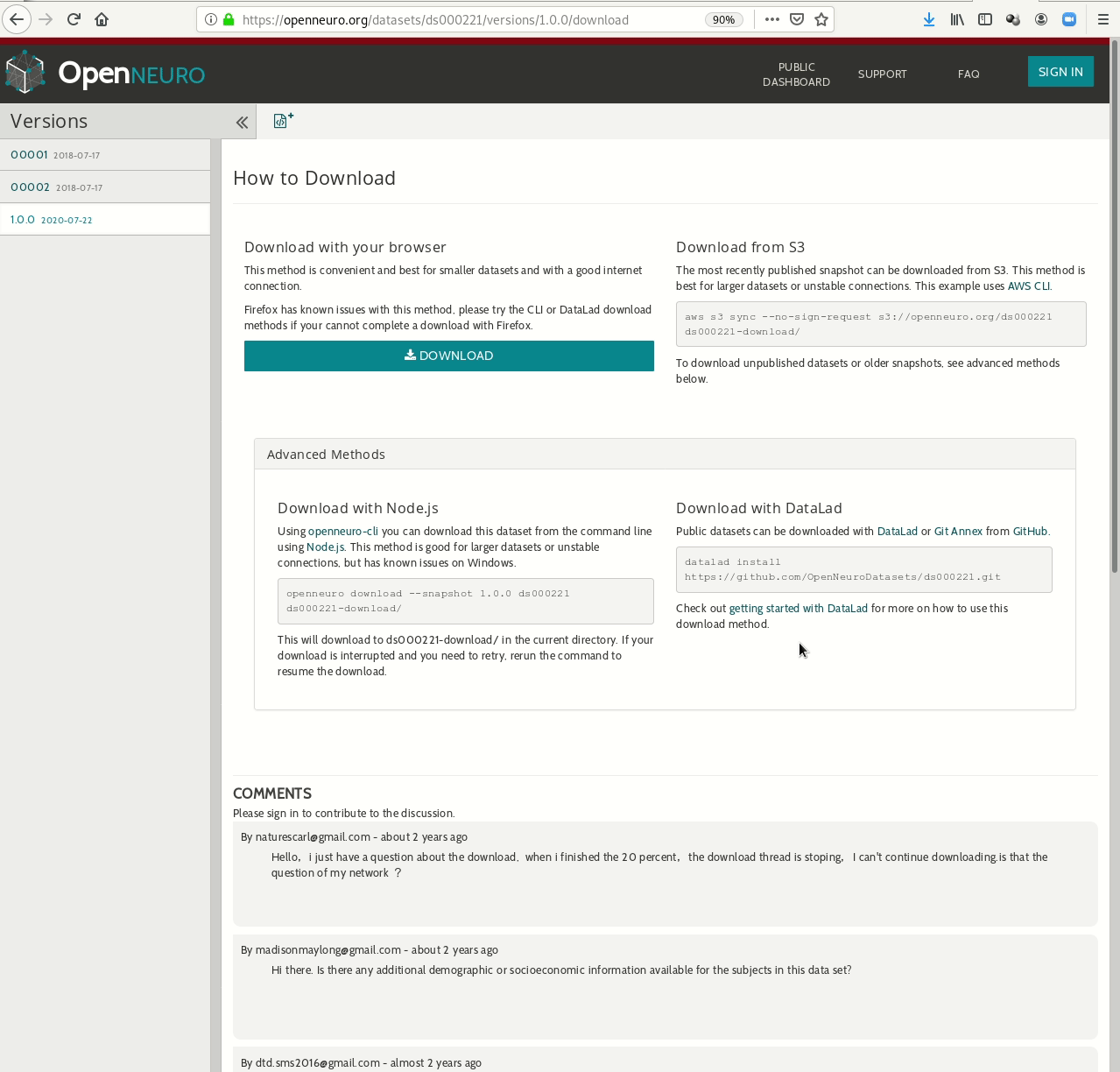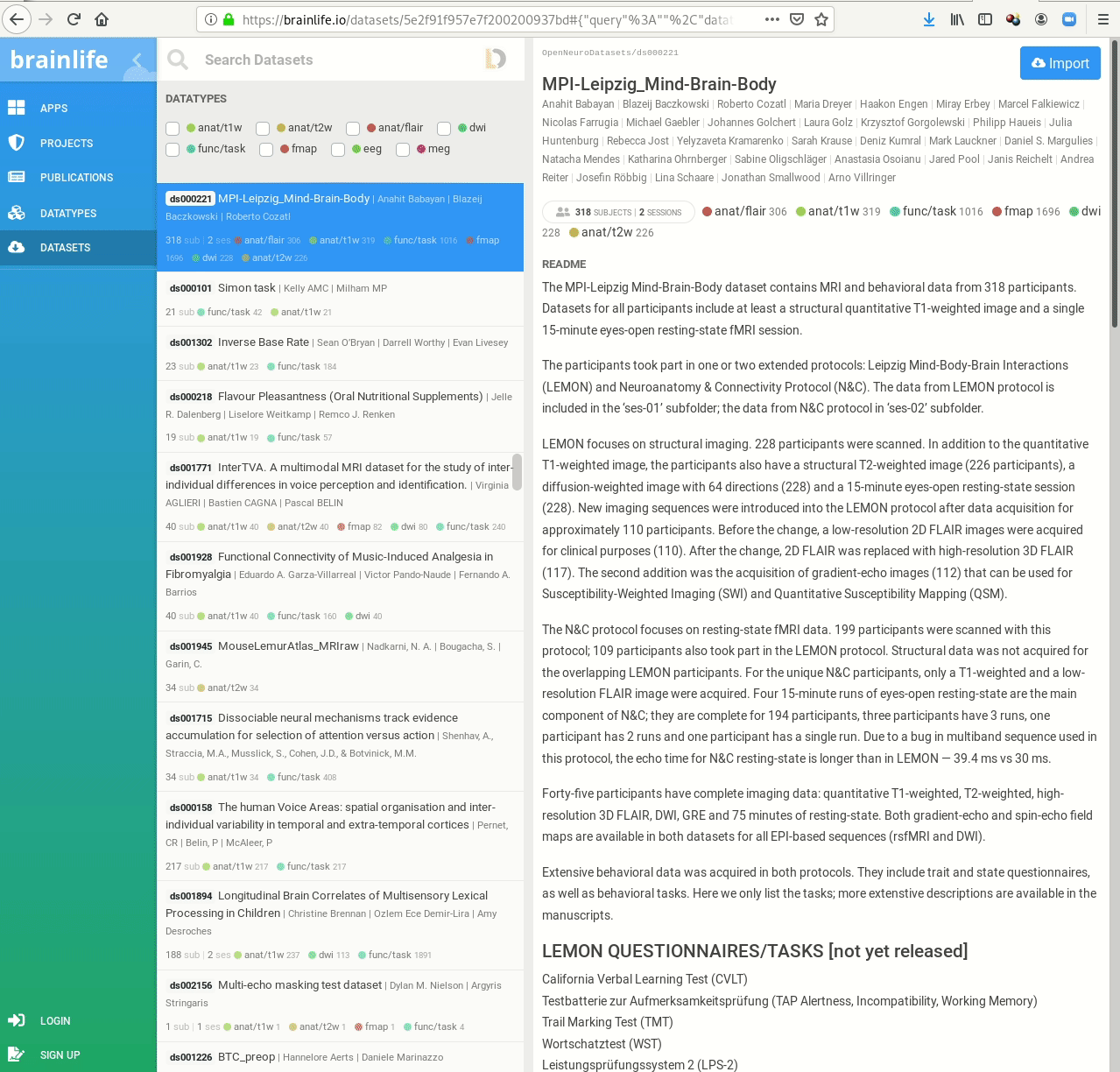
Examples of what DataLad can be used for:
- Central data management and archival system
Examples of what DataLad can be used for:
-
... and much more!
Code along
Code to follow along: handbook.datalad.org/en/latest/code_from_chapters/MPI_code.htmlVersion control
Why version control?
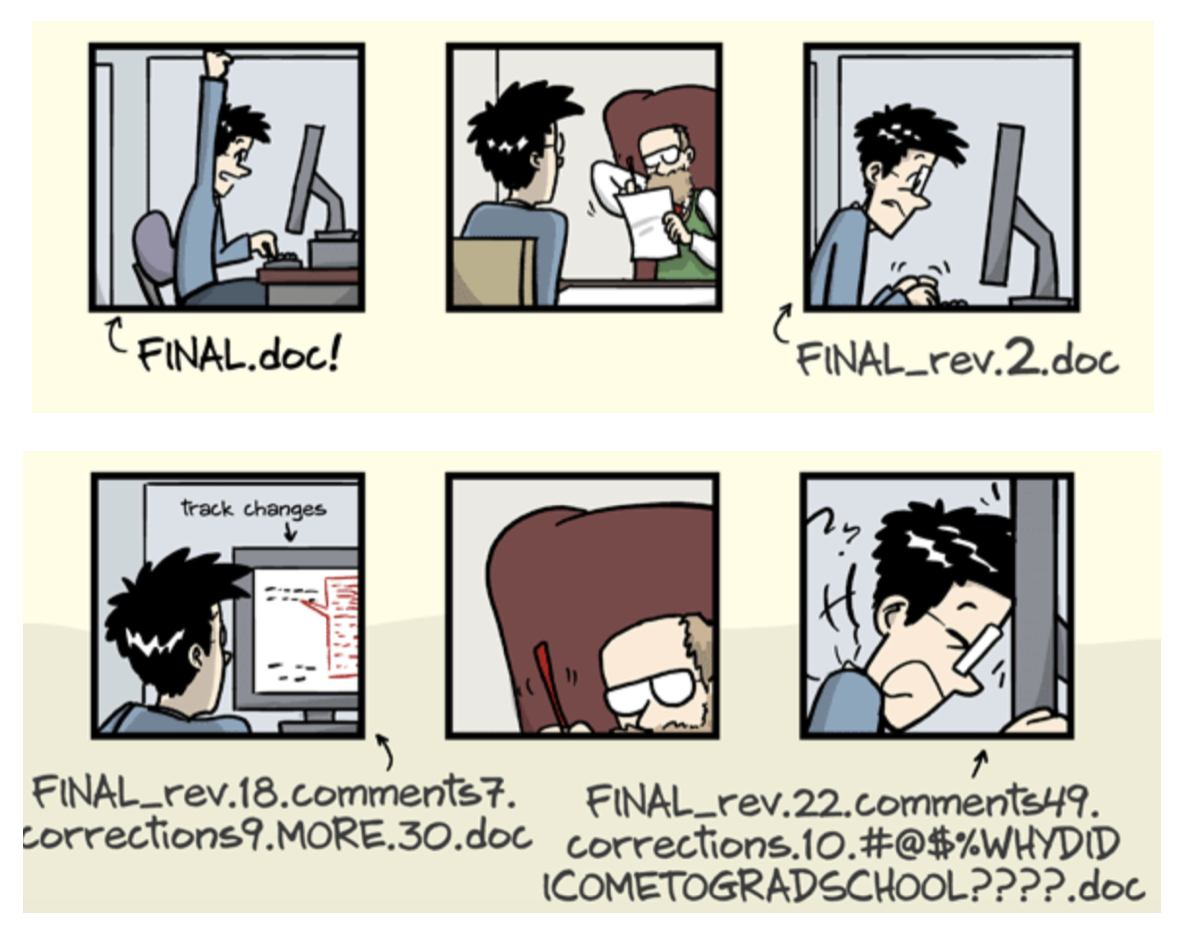
- keep things organized
- keep track of changes
- revert changes or go back to previous states
Version Control
- DataLad knows two things: Datasets and files
- A DataLad dataset is a Git/git-annex: repository:
- For Git users: Use workflows from software development for science!
- Content and domain agnostic - Manage science, or your music library
- Minimization of custom procedures or data structures - A PDF stays a PDF, and users won't lose data or data access if DataLad vanishes
Local version control
Procedurally, version control is easy with DataLad!
Advice:
- Save meaningful units of change
- Attach helpful commit messages
Summary - Local version control
datalad createcreates an empty dataset.- Configurations (-c yoda, -c text2git) are useful (details soon).
- A dataset has a history to track files and their modifications.
- Explore it with Git (git log) or external tools (e.g., tig).
datalad saverecords the dataset or file state to the history.- Concise commit messages should summarize the change for future you and others.
datalad download-urlobtains web content and records its origin.- It even takes care of saving the change.
datalad statusreports the current state of the dataset.- A clean dataset status is good practice.
Questions!
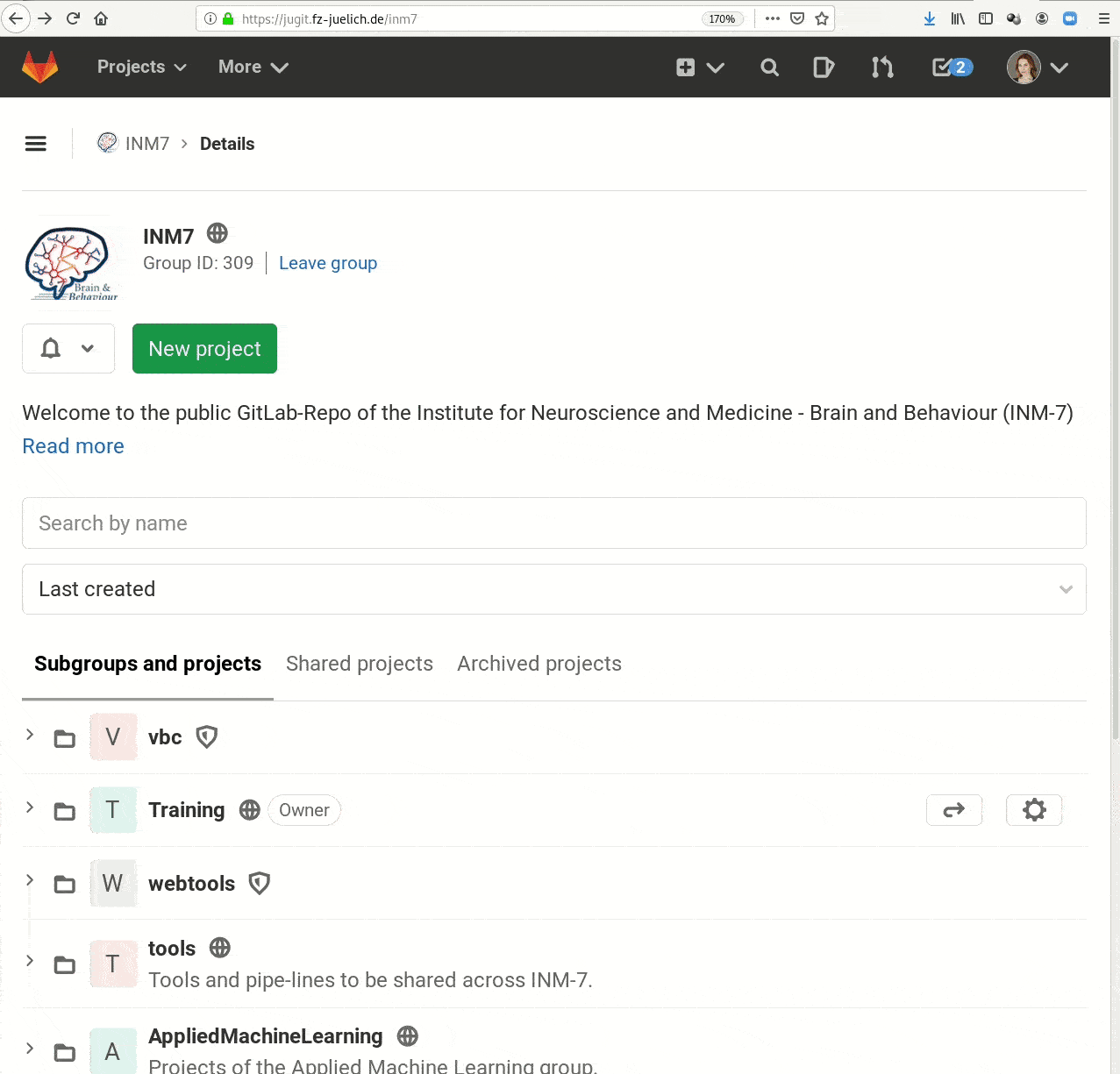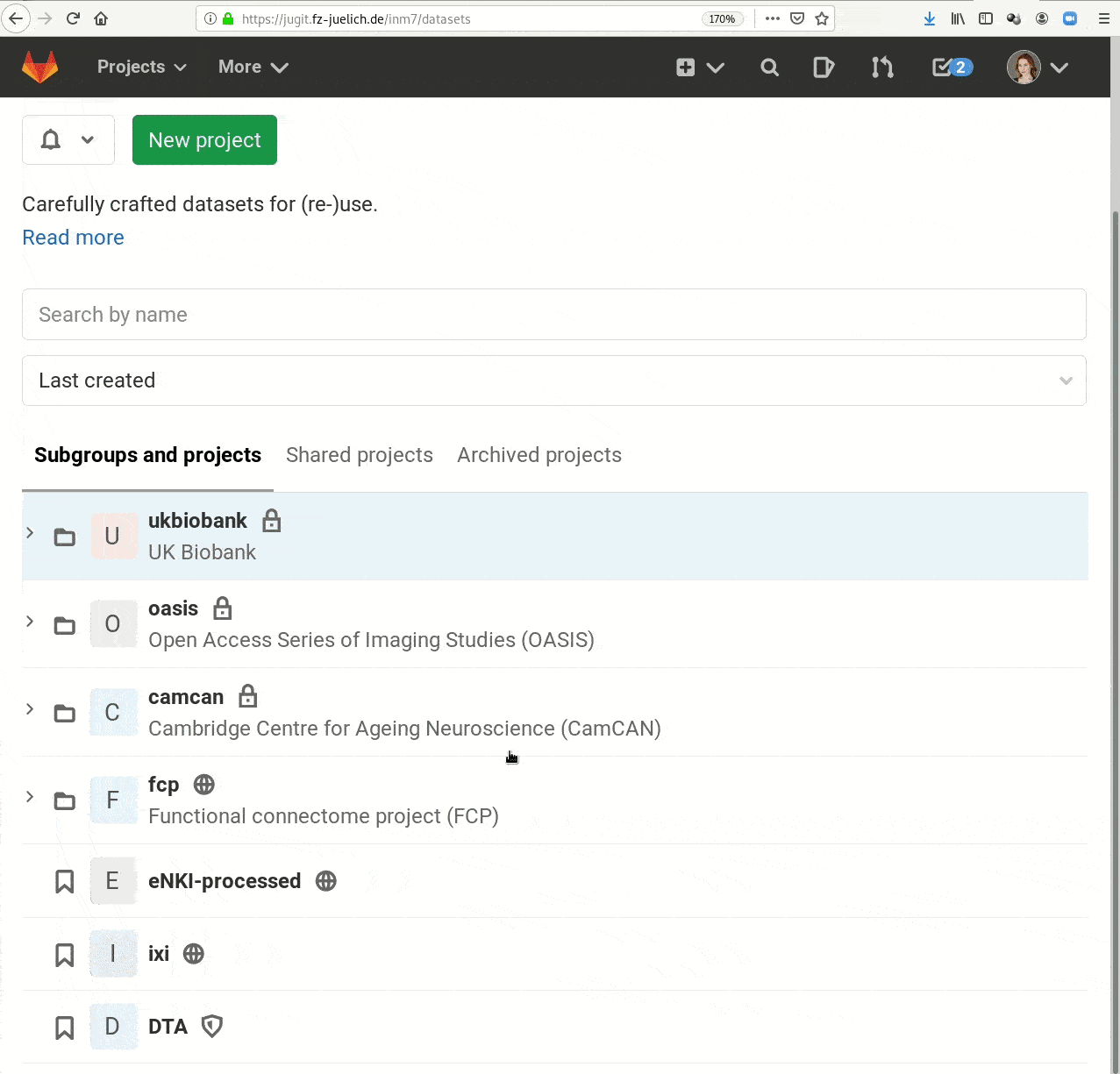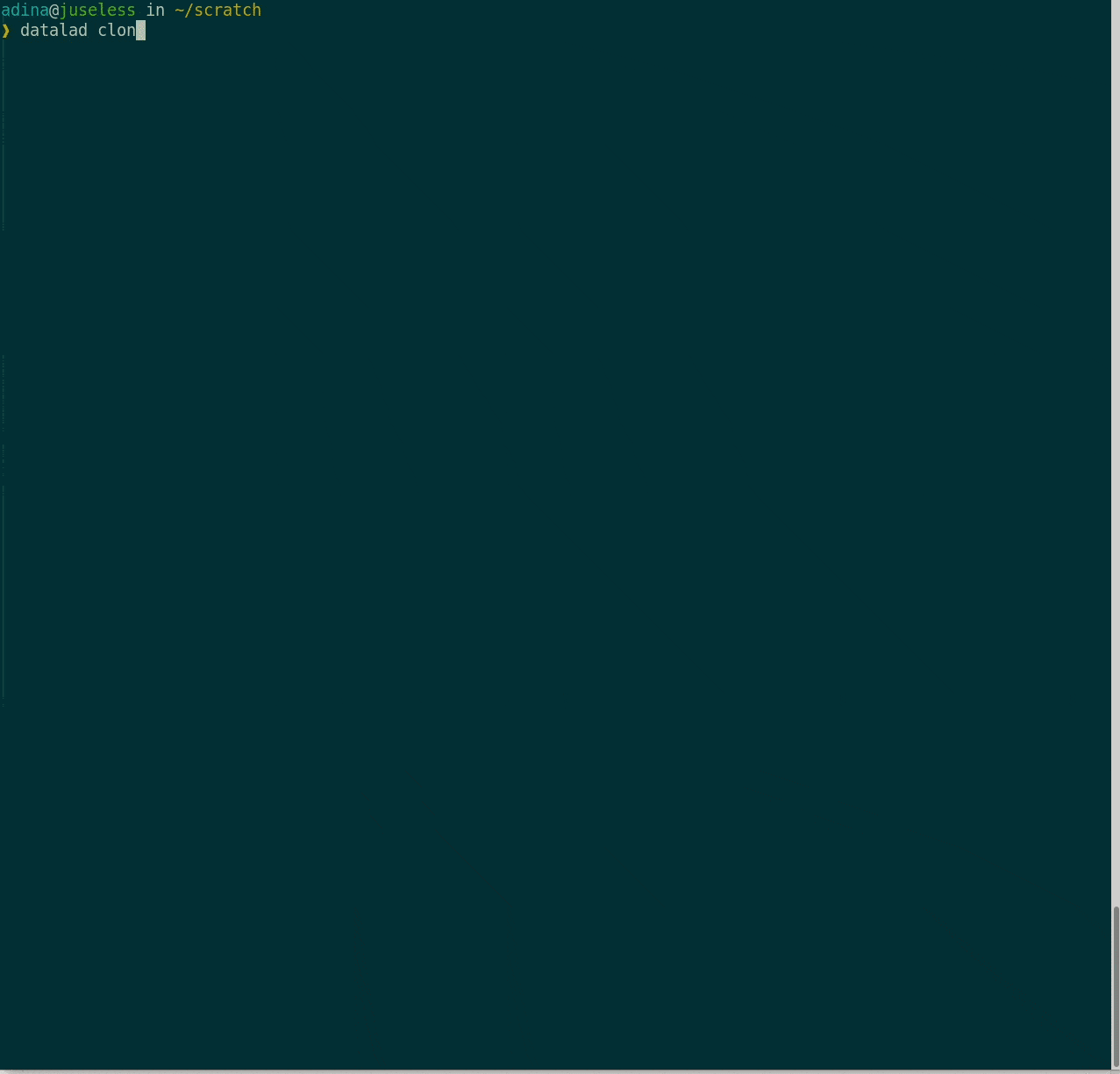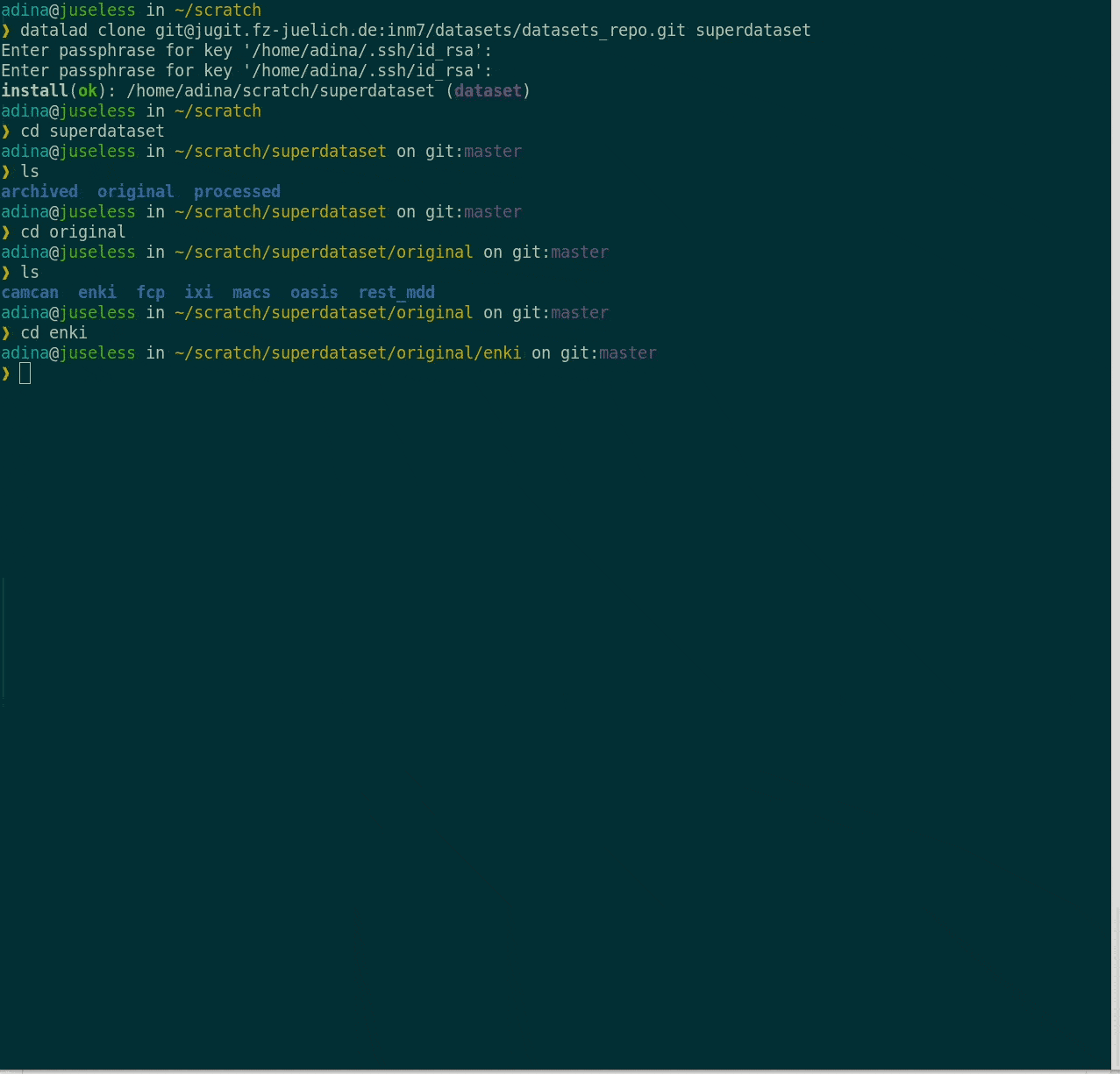
Consuming datasets
- Here's how a dataset looks after installation:
Plenty of data, but little disk-usage
- Cloned datasets are lean. "Meta data" (file names, availability) are present, but no file content:
$ datalad clone git@github.com:psychoinformatics-de/studyforrest-data-phase2.git
install(ok): /tmp/studyforrest-data-phase2 (dataset)
$ cd studyforrest-data-phase2 && du -sh
18M .$ ls
code/
src/
stimuli
sub-01/
sub-02/
sub-03/
sub-04/
[...]$ datalad get sub-01/ses-movie/func/sub-01_ses-movie_task-movie_run-1_bold.nii.gz
get(ok): /tmp/studyforrest-data-phase2/sub-01/ses-movie/func/sub-01_ses-movie_task-movie_run-1_bold.nii.gz (file) [from mddatasrc...]# eNKI dataset (1.5TB, 34k files):
$ du -sh
1.5G .
# HCP dataset (80TB, 15 million files)
$ du -sh
48G .
Sharing datasets
- Share data with others and keep them up to date, or get data from
someone and stay up to date (
datalad update --merge) - Have all updates in your dataset history, but pick the version you want to work with
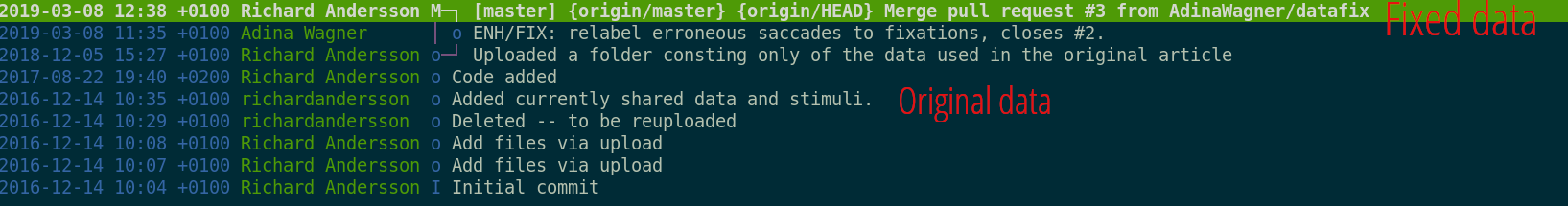
Summary - Dataset consumption & nesting
datalad cloneinstalls a dataset.- from local or remote sources.
- Datasets can be installed as subdatasets within an existing dataset.
- Using the --dataset/-d option. Useful for transparency, cleanliness, and scalability.
- Only small files and file availability metadata are present.
datalad getretrieves file contents on demand,datalad dropcan remove file content on demand.- Datasets preserve their history.
- Superdatasets record the version state of their subdataset.
Questions!
reproducible data analysis
Your past self is the worst collaborator:
Basic organizational principles for datasets
- Keep everything clean and modular
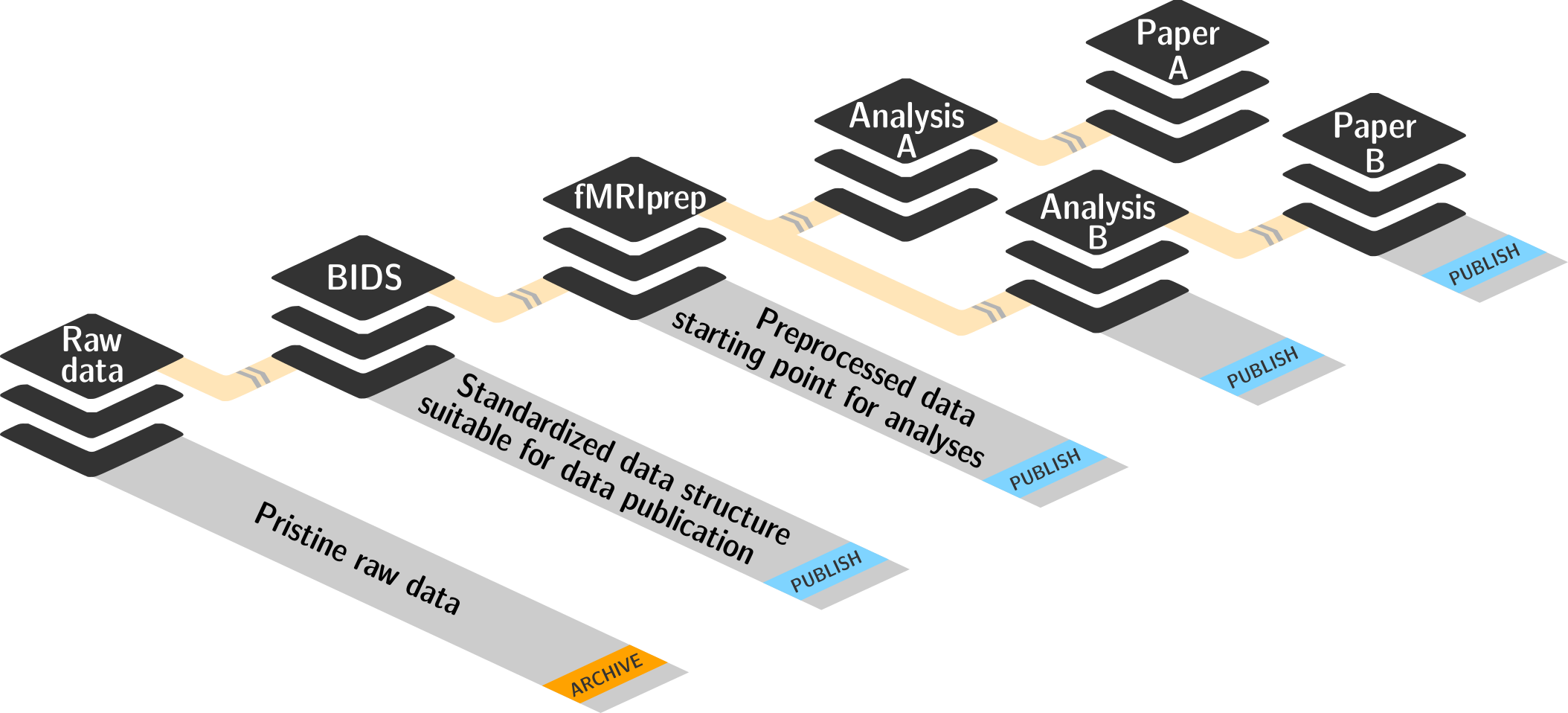 |
|
- do not touch/modify raw data: save any results/computations outside of input datasets
- Keep a superdataset self-contained: Scripts reference subdatasets or files with relative paths
Basic organizational principles for datasets
- Record where you got it from, where it is now, and what you do to it
- Document everything:
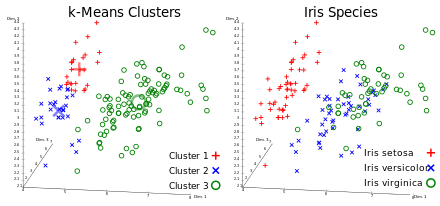
A classification analysis on the iris flower dataset
Reproducible execution & provenance capture
datalad run
Provenance capture
- Those "run records" are stored in a dataset's history and can be automatically rerun:
$ datalad rerun eee1356bb7e8f921174e404c6df6aadcc1f158f0
[INFO] == Command start (output follows) =====
[INFO] == Command exit (modification check follows) =====
add(ok): sub-01/LC_timeseries_run-1.csv (file)
...
save(ok): . (dataset)
action summary:
add (ok: 45)
save (notneeded: 45, ok: 1)
unlock (notneeded: 45)
...Computational reproducibility
- Code may fail (to reproduce) if run with different software
- Datasets can store (and share) software environments (Docker or Singularity containers) and reproducibly execute code inside of the software container, capturing software as additional provenance
- DataLad extension:
datalad-container
datalad-containers run
Summary - Reproducible execution
datalad runrecords a command and its impact on the dataset.- All dataset modifications are saved - use it in a clean dataset.
- Data/directories specified as
--inputare retrieved prior to command execution. - Use one flag per input.
- Data/directories specified as
--outputwill be unlocked for modifications prior to a rerun of the command. - Its optional to specify, but helpful for recomputations.
datalad containers-runcan be used to capture the software environment as provenance.- Its ensures computations are ran in the desired software set up.
datalad reruncan automatically re-execute run-records later.- They can be identified with any commit-ish (hash, tag, range, ...)
Questions!
Interested in more about computational reproducibility? Checkout the usecase DataLad for machine-learning anlaysis at handbook.datalad.orgDatasets for yourself and others
- DataLad is built to maximize interoperability and use with hosting and storage technology: Share datasets with the services you use anyway
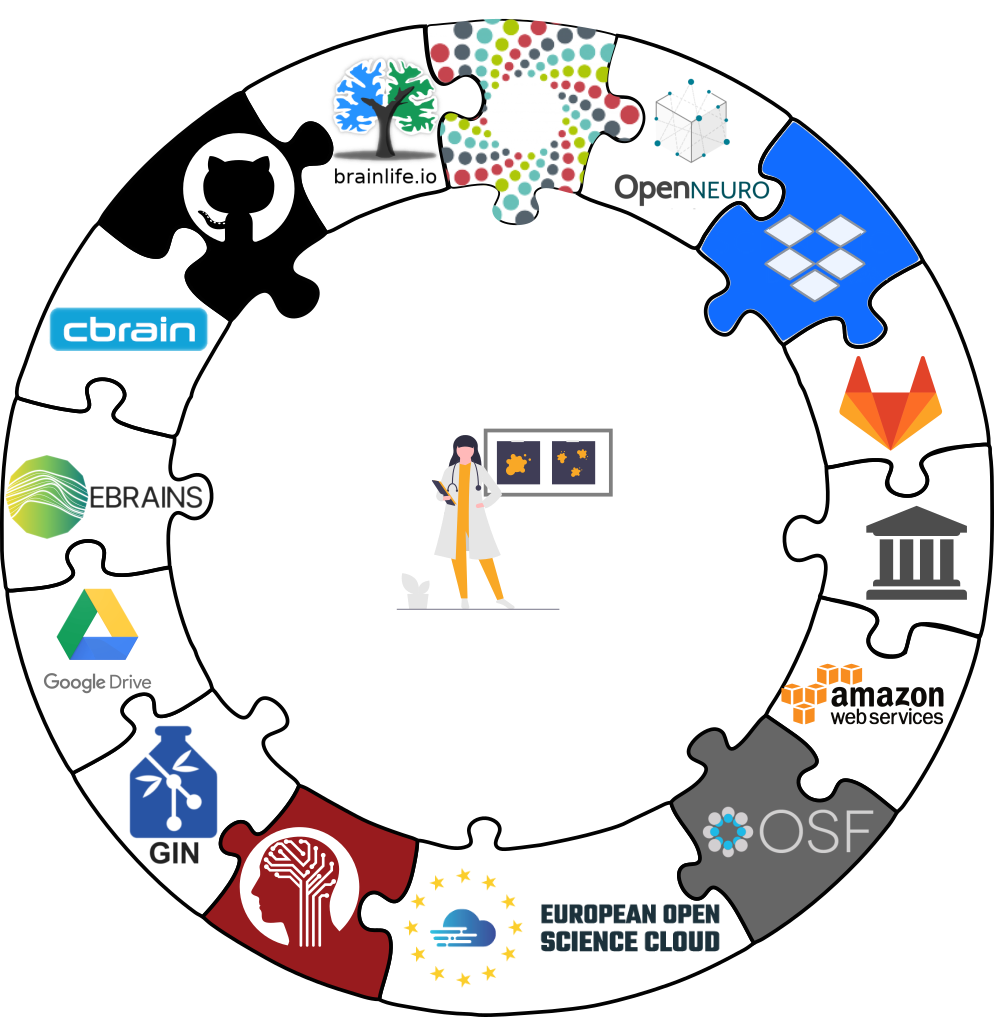
Datasets for yourself and others
- DataLad is built to maximize interoperability and use with hosting and storage technology: Share datasets with the services you use anyway
Everything you need to know about sharing datasets is in the chapter in Third party infrastructure
Why use DataLad?
- Mistakes are not forever anymore: Easy version control, regardless of file size
- Who needs short-term memory when you can have run-records?
- Disk-usage magic: Have access to more data than your hard drive has space
- Collaboration and updating mechanisms: Alice shares her data with Bob. Alice fixes a mistake and pushes the fix. Bob says "datalad update" and gets her changes. And vice-versa.
- Transparency: Shared datasets keep their history. No need to track down a former student, ask their project what was done.
- No need to ask colleagues what they did, you can ask the files how they came to be:
$ git log some_result_file
commit 593aa8018116ca9d198ce4bfd9e09af3476c7a9b
Author: Elena Piscopia elena@example.net
Date: Thu Sep 3 13:35:51 2020 +0200
[DATALAD RUNCMD] Re-create the results with most recent data
=== Do not change lines below ===
{
"chain": [
"38e18c0cd73627e10b620b1ba08e4be2caba18e7"
],
"cmd": "bash code/mycode.sh",
"dsid": "57ce4457-a29b-4bd0-be6f-a9da8d46aee3",
"exit": 0,
"extra_inputs": [],
"inputs": data/input_data/*.nii.gz,
"outputs": [],
"pwd": "."
}
^^^ Do not change lines above ^^^
$ datalad rerun 593aa8018116caQuestions!
Real-life examples
(Raw) data mismanagement
- Multiple large datasets are available on a compute cluster 🏞
- Each researcher creates their own copies of data ⛰
- Multiple different derivatives and results are computed from it 🏔
- Data, copies of data, half-baked data transformations, results, and old versions of results are kept - undocumented 🌋
Example: eNKI dataset
- Raw data size: 1.5 TB
- + Back-up: 1.5 TB
- + A BIDS structured version: 1.5 TB
- + Common, minimal derivatives (fMRIprep): ~ 4.3TB
- + Some other derivatives: "Some other" x 5TB
- + Copies of it all or of subsets in home and project directories
Example: eNKI dataset



"Can't we buy more hard drives?"









No.







DataLad way
- Download the data, have a back-up
- Transform it into a DataLad dataset
$ datalad create -f .
$ datalad save -m "Snapshot raw data"$ datalad create my_enki_analysis
$ datalad clone -d . /data/enki dataLack of provenance can be devastating
- Data analyses typically start with data wrangling:
- Move/Copy/Rename/Reorganize/... data
- Mistakes propagate through the complete analysis pipeline - especially those early ones are hard to find!
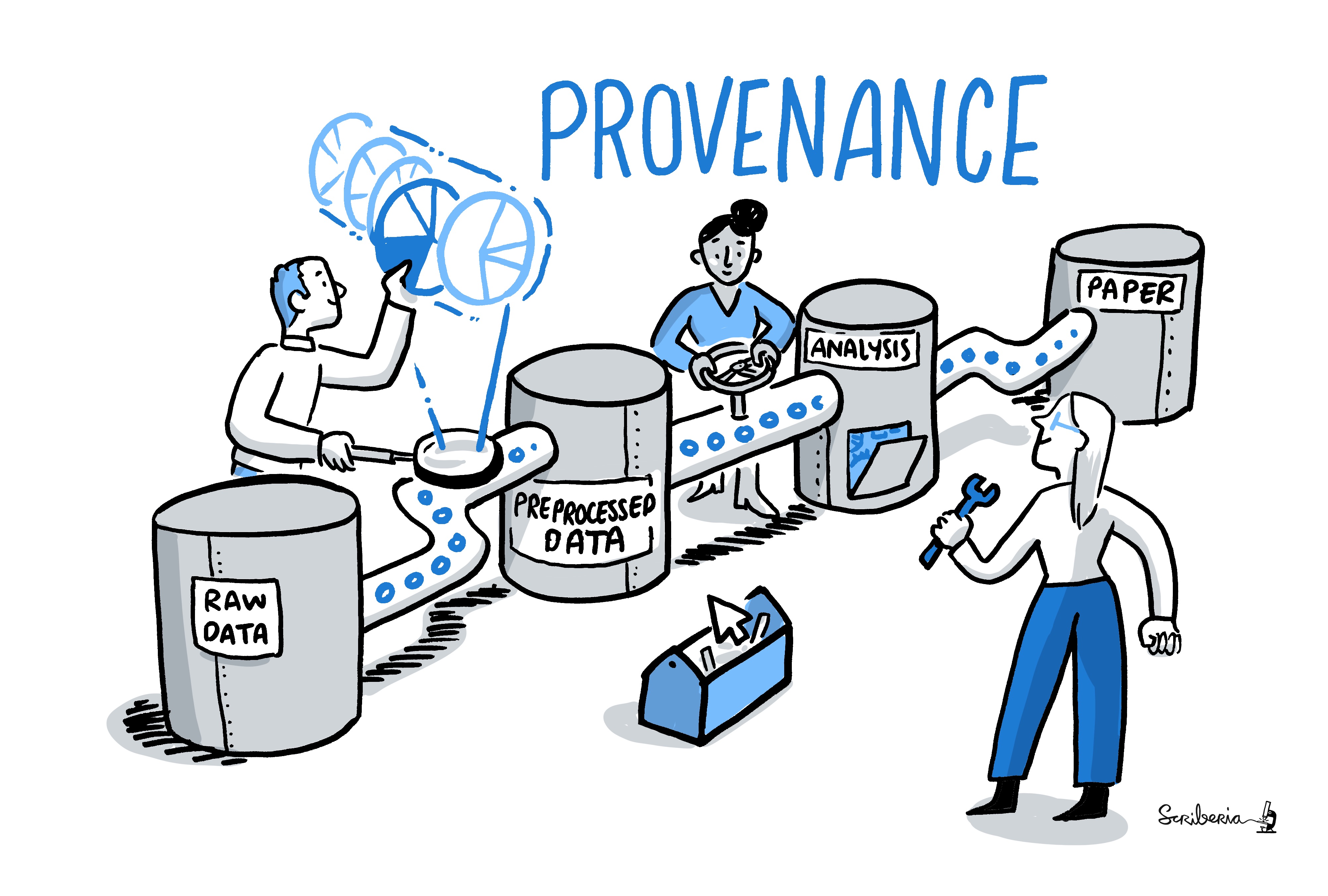
Example: "Let me just copy those files..."
- Researcher builds an analysis dataset and moves
events.tsvfiles (different per subject) to the directory with functional MRI data
$ for sourcefile, dest in zip(glob(path_to_events), # note: not sorted!
glob(path_to_fMRI_subjects)): # note: not sorted!
destination = path.join(dest, Path(sourcefile).name)
shutil.move(sourcefile, destination)Researcher shares analysis with others
😱
- organized
- knowledgeable
- experienced

Everyone makes mistakes - the earlier we find them or guard against them, the better for science!
Leave a trace!
$ datalad run -m "Copy event files" \
"for sub in eventfiles;
do mv ${sub}/events.tsv analysis/${sub}/events.tsv;
done"
$ datalad copy-file ../eventfiles/sub-01/events.tsv sub-01/ -d .
copy_file(ok): /data/project/coolstudy/eventfiles/events.tsv [/data/project/coolstudy/analysis/sub-01/events.tsv]
save(ok): /data/project/coolstudy/analysis (dataset)
action summary:
copy_file (ok: 1)
save (ok: 1)Writing a reproducible paper
Live-Demo!- GitHub repository: github.com/psychoinformatics-de/paper-remodnav
- Detailed write-up and tutorial: handbook.datalad.org/en/latest/usecases/reproducible-paper.html
Writing a reproducible paper
- The details of how the reproducible paper was created (Makefiles, Python code, LaTeX-based manuscript) are arbitrary - there are many ways of creating them.
- What I regard as important is the backbone that DataLad provides: A vehicle to link data to code and distribute it alongside to it and means to collaboratively work on science as one would in software development
Thank you!
Back-up/Details
Git versus Git-annex
- Data in datasets is either stored in Git or git-annex
- By default, everything is stored in git-annex
| Git | git-annex |
| handles small files well (text, code) | handles all types and sizes of files well |
| file contents are in the Git history and will be shared upon git/datalad push | file contents are in the annex. Not necessarily shared |
| Shared with every dataset clone | Can be kept private on a per-file level when sharing the dataset |
| Useful: Small, non-binary, frequently modified, need-to-be-accessible (DUA, README) files | Useful: Large files, private files |
Git versus Git-annex
Useful background information for demo later. Read this handbook chapter for detailsGit and Git-annex handle files differently: annexed files are stored in an annex. File content is hashed & only content-identity is committed to Git.
- Files stored in Git are modifiable, files stored in Git-annex are content-locked
- Annexed contents are not available right after cloning, only content- and availability information (as they are stored in Git)
|
|
|
Git versus Git-annex
-
When sharing datasets with someone without access to the same computational
infrastructure, annexed data is not necessarily stored together with the rest
of the dataset.

-
Transport logistics exist to interface with all major storage providers.
If the one you use isn't supported, let us know!
Git versus Git-annex
-
Users can decide which files are annexed:
- Pre-made run-procedures, provided by DataLad (e.g.,
text2git,yoda) or created and shared by users (Tutorial at handbook.datalad.org) - Self-made configurations in
.gitattributes(e.g., based on file type, file/path name, size, ...) - Per-command basis (e.g., via
datalad save --to-git)
Datasets scale!
adina@bulk1 in /ds/hcp/super on git:master❱ datalad status --annex -r
15530572 annex'd files (77.9 TB recorded total size)
nothing to save, working tree cleanFind out more
|
Comprehensive user documentation in the DataLad Handbook (handbook.datalad.org) |
|
|
|
|
|
|
Further info and reading
Everything I am talking about is documented in depth elsewhere:- General DataLad tutorial: handbook.datalad.org/basics/intro.html/
- How to structure data analysis projects: handbook.datalad.org/r.html?yoda
- More DataLad tutorials: DataLad YouTube channel
Open an issue on GitHub if you have more questions!
