Research Data Management for big data
🚀
DataLad and the Human Connectome Project
Adina Wagner
 @AdinaKrik
@AdinaKrik |
|
|
Psychoinformatics lab,
Institute of Neuroscience and Medicine, Brain & Behavior (INM-7) Research Center Jülich |
Slides: https://github.com/datalad-handbook/course/
Research data management (RDM)
- (Research) Data = every digital object involved in your project: code, software/tools, raw data, processed data, results, manuscripts ...
- Data needs to be managed FAIRly- from creation to use, publication, sharing, archiving, re-use, or destruction:

- Research data management is a key component for reproducibility, efficiency, and impact/reach of data analysis projects
Why data management?

⬆
This a metaphor for most projects after publication
Why data management?
This a metaphor for reproducing (your own) research
a few months after publication
⬇

Why data management?
| This is a metaphor for many computational ➡ clusters without RDM |
 |
Why data management?
- External requirements and expectations
- Funders & publishers require it
- Scientific peers increasingly expect it
- Intrinsic motivation and personal & scientific benefits
- The quality, efficiency and replicability of your work improves
- The most interesting datasets of our field require it
- Exciting datasets (UKBiobank, HCP, ...) are orders of magnitudes larger than previous public datasets, and neither the computational infrastructure nor analysis workflows scale to these dataset sizes
A common tale of RDM in science
- Multiple large datasets are available on a compute cluster 🏞
- Each researcher creates their own copies of data ⛰
- Multiple different derivatives and results are computed from it 🏔
- Data, copies of data, half-baked data transformations, results, and old versions of results are kept - undocumented 🌋
Example: eNKI dataset

- Raw data size: 1.5 TB
- + Back-up: 1.5 TB
- + A BIDS structured version: 1.5 TB
- + Common, minimal derivatives (fMRIprep): ~ 4.3TB
- + Some other derivatives: "Some other" x 5TB
- + Copies of it all or of subsets in home and project directories
How much storage capacity does a typical compute cluster have?


Can we buy more hard drives?









Depends







If your institution doesn't care about money or the
environment, more disk space can help...
💸🤷🌏
But with a certain amount of data, simply "stocking up"
becomes not only ridiculous, but also infeasible:
HCP: 80TB
UKBiobank (current): 42TB
- Introduction and core concepts
- Hands-on: How can I use it with the HCP data?
Acknowledgements
|
Funders






Collaborators





|
 Core Features:
Core Features:
- Joint version control (Git, git-annex) for code, software, and data
- Provenance capture: Create and share machine-readable, re-executable records of your data analysis for reproducible, transparent, and FAIR research
- Data transport mechanisms: Install or share data extremely lightweight, retrieve it on demand, drop it to free up space without losing data access or provenance
Examples of what DataLad can be used for:
- Publishing datasets and making them available via GitHub, GitLab, or similar services
- Creating and sharing reproducible, open science: Sharing data, software, code, and provenance
- Behind-the-scenes infrastructure component for data transport and versioning (e.g., used by OpenNeuro, brainlife.io , the Canadian Open Neuroscience Platform (CONP), CBRAIN)
- Central data management and archival system (pioneered at the INM-7, Research Centre Juelich)
Find out more
|
Comprehensive user documentation in the DataLad Handbook (handbook.datalad.org) |
|
|
|
|
|
|
Requirements
- DataLad version 0.12.2 or later (Installation instructions at handbook.datalad.org)
- Human Connectome Project AWS credentials (register at db.humanconnectome.org/, enable S3 Access)
Further info and reading
Everything I am talking about is documented in depth elsewhere:- General DataLad tutorial: handbook.datalad.org/basics/intro.html/
- Info on the HCP DataLad dataset: handbook.datalad.org/usecases/HCP_dataset.html
- Instructions & example on processing large dataset with HTCondor: handbook.datalad.org/beyond_basics/101-170-dataladrun.html
- How to structure data analysis projects: handbook.datalad.org/r.html?yoda
HCP data structure
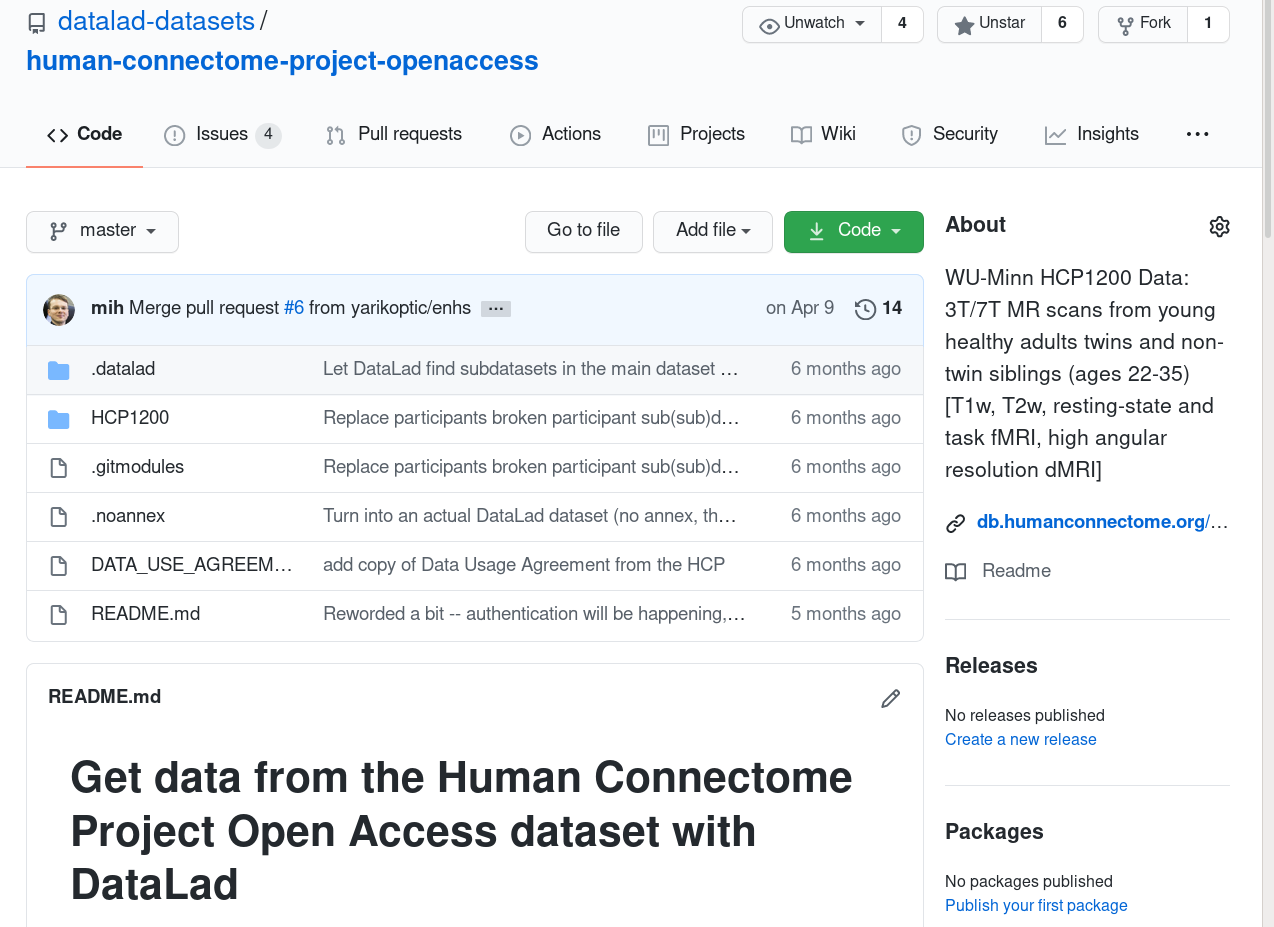
- HCP data is available in full, or in subsets (for speedier installation), from GitHub:
Get HCP data via DataLad
- datalad clone a GitHub repository
$ datalad clone \
git@github.com:datalad-datasets/human-connectome-project-openaccess.git \
HCP
install(ok): /.../HCP (dataset)$ datalad get HCP1200/221218/T1w/T1w_acpc_dc.nii.gz
install(ok): /tmp/HCP/HCP1200/221218/T1w (dataset)
[Installed subdataset in order to get /tmp/HCP/HCP1200/221218/T1w/T1w_acpc_dc.nii.gz]
get(ok): /tmp/HCP/HCP1200/221218/T1w/T1w_acpc_dc.nii.gz (file) [from datalad...]
action summary:
get (ok: 1)
install (ok: 1)HCP dataset structures
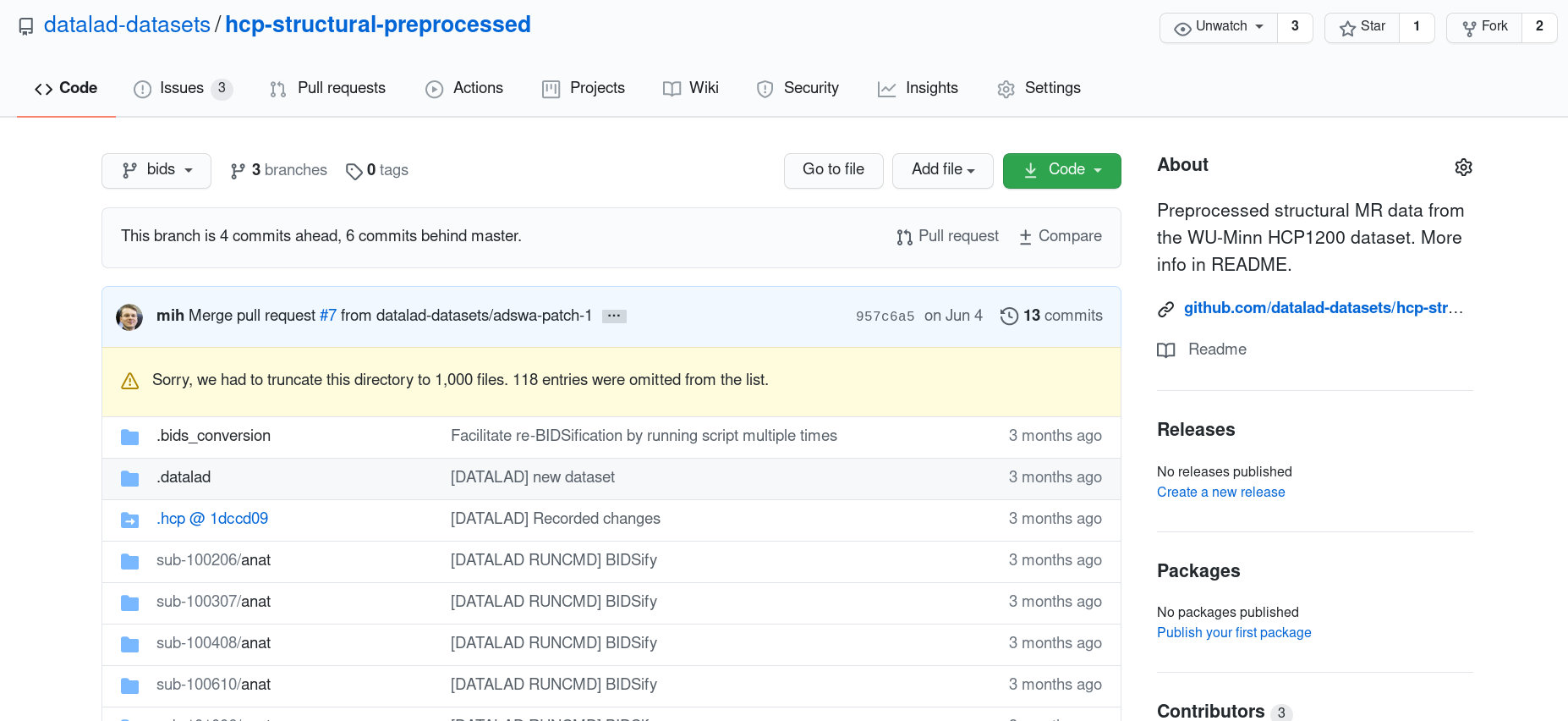
- Dataset structure follows HCP data layout:
- subject-ID
- data directories (unprocessed, T1w, MNINonLinear, MEG, release notes)
- The full HCP dataset consists of numerous subdatasets (subjects, data directories)
HCP dataset structures
- The HCP subset datasets usually are a single dataset (advantage: much faster installation)
- Some are available in BIDS-like structures
- You can create such subsets yourself or request them by emailing us paths
FAIR, large-scale data processing
Basic organizational principles for datasets
Read all about this in the chapter on YODA principles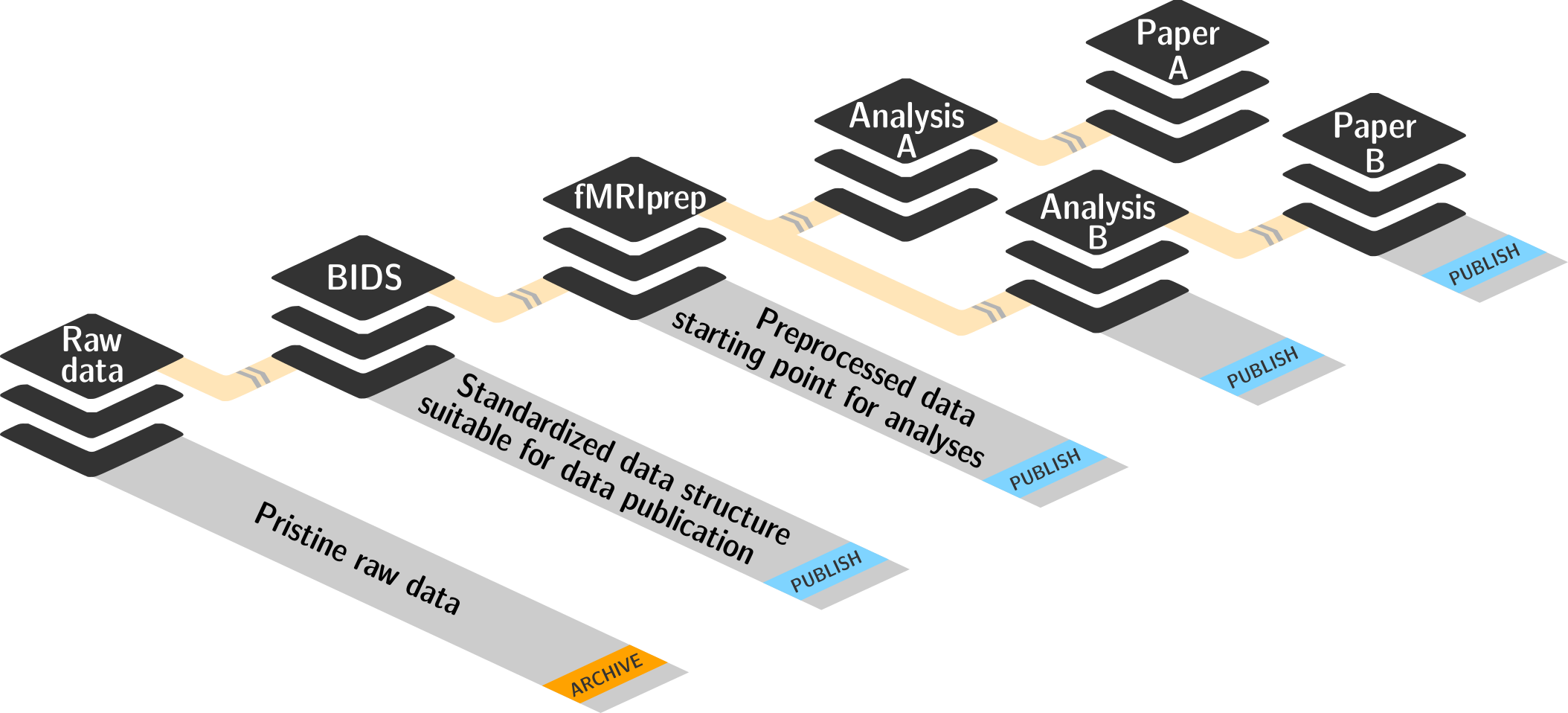 |
|
- do not touch/modify raw data: save any results/computations outside of input datasets
- Keep a superdataset self-contained: Scripts reference subdatasets or files with relative paths
Basic organizational principles for datasets
- Record where you got it from, where it is now, and what you do to it
